Help
From GreeNC
Contents
About GreeNC
Over the last few years, it has emerged the idea that long non-coding RNAs (lncRNAs) might have a very important role in transcriptional regulation and control of gene expression. The Green Non-Coding database (GeeNC) was born in 2014 with the aim to provide a comprehensive annotation of lncRNAs among different plant species to the research community.
GreeNC is the major resource of plant lncRNAs today with more than 200,000 transcripts annotated with a wide range of information about them. This database has future perspectives of adding new species and de-novo annotated transcripts with RNA-seq data besides of carrying out phylogenetic studies about this class of non-coding RNAs.
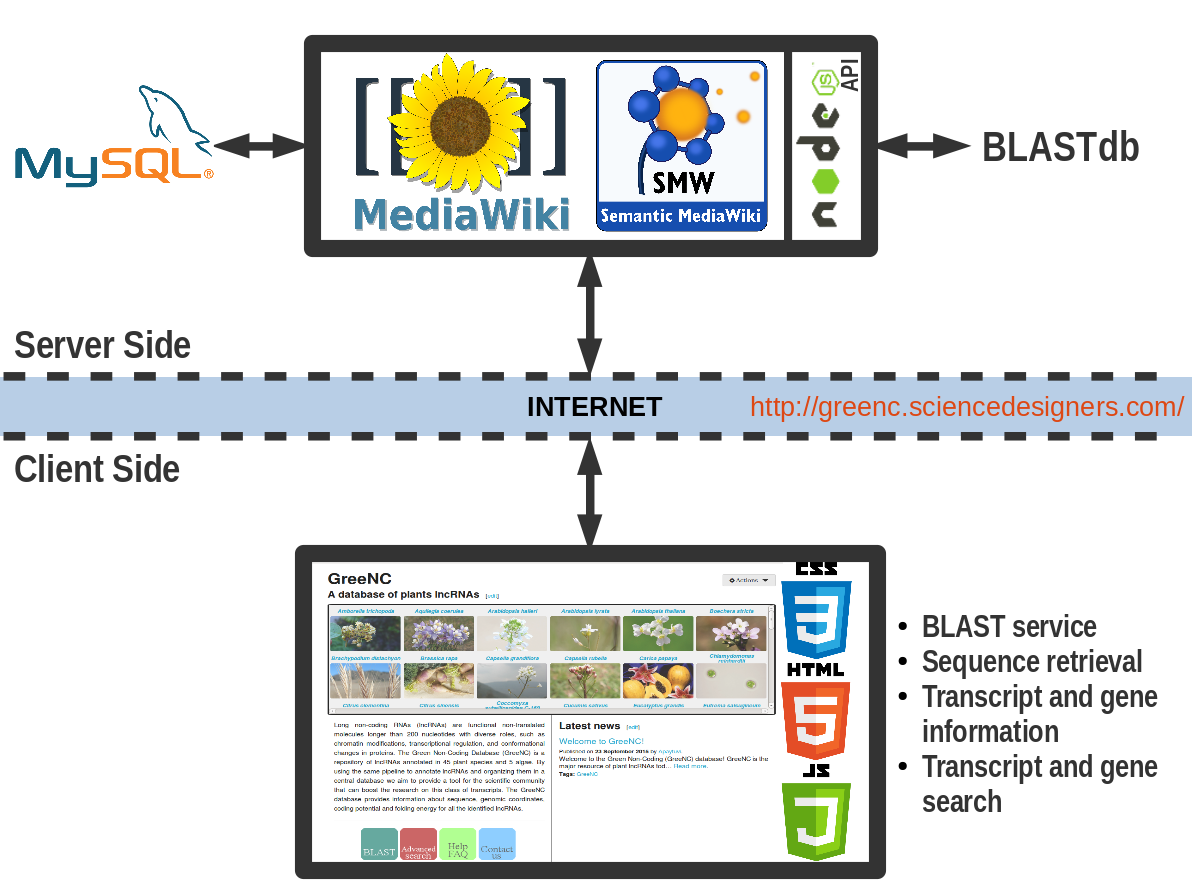
This database is based on MySQL and has information about transcript and gene features of lncRNAs. The data is integrated into a MediaWiki by mapping relational data fields against wiki predefined templates via Semantic MediaWiki. All transcript sequences are kept in a FASTA file with the same IDs as kept in MySQL and then formatted using NCBI makeblastdb. Taking advantage of this, an Express NodeJS API webservice was created in order to expose both sequence retrieval and BLAST searches via client JavaScript from the MediaWiki interface (Figure 1).
Gene and transcript aliases of GreeNC have the following structre: short-species-cientific-name_gene-or-transcript-name. For example, the gene AT1G01170 and the transcript AT1G01170.1 would have in GreeNC the corresponding aliases: Athaliana_AT1G01170 and Athaliana_AT1G01170.1.
Each gene page displays information about the locus and its non-coding transcripts using two tables called Gene information and Transcript features. If there exists any hit/association/coincidence with an external database (SwissProt, miRBase, Rfam, RepBase, NONCODE, or lncRNAdb), a third table called Matches to external databases will be displayed.
How does GreeNC differ from other databases?
All lncRNAs from GeeNC have been annotated in silico from reference transcripts using highly specific and sensitive in-house bioinformatics pipelines (look at What is the criteria for a lncRNA being added to GreeNC?). We have identified putative lncRNAs of 50 species using Phytozome v10.3 annotations.
The fact that GreeNC does not focus on just one species, but focuses on as many plant species as possible, makes this database have a cross-sectional character, being highly attractive to the plant research community.
What is the criteria for a lncRNA being added to GreeNC?
The lncRNAs contained by GreeNC have the following common features:
- They are larger than 200 nt.
- They have an open reading frame (ORF) smaller than 360 nt (120 aa).
- Either they do not have a hit to SwissProt or they are classified as non-coding by the Coding Potential Calculator (CPC).
- They are not classified either as rRNA, tRNA, snRNA, nor snoRNA by hmmer against Rfam.
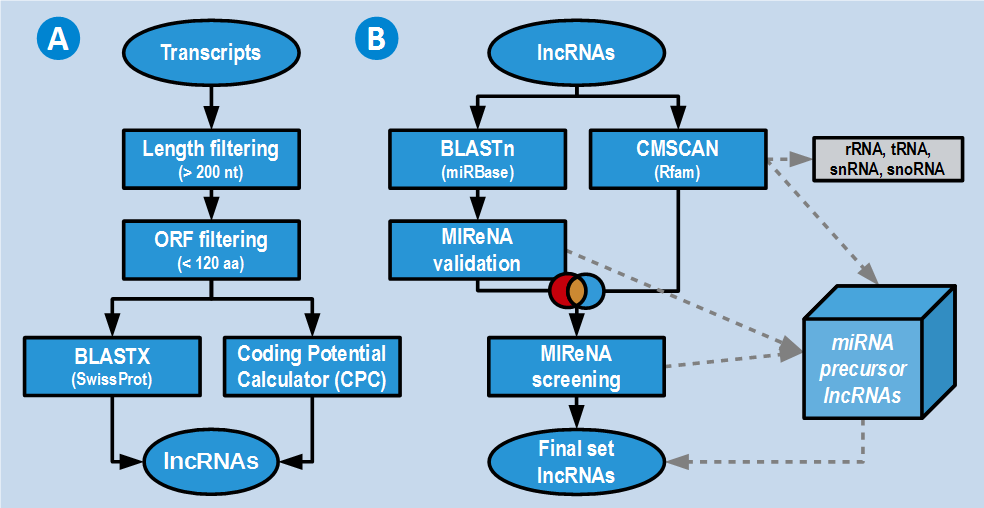
These features have been assessed using highly specific and sensitive in-house bioinformatics pipelines. First 3 features have been assessed by script 1 (Figure 2A). Last feature has been assessed by script 2 (Figure 2B).
Those transcripts 1) without hits in SwissProt, 2) described as non-coding by the CPC, and 3) considered non-precursors of miRNA are classified as high-confidence lncRNAs. Transcripts without hits in SwissProt but described as coding by CPC or transcripts with hits in SwissProt but described as non-coding by CPC are considered low-confidence lncRNAs. Those transcripts identified as putative precursors of miRNAs or having along their sequence repetitive elements predicted by RepeatMasker using Repbase are also considered low-confidence lncRNAs.
Is it possible to perform bulk downloads?
Indeed. You can download all sequences for each species in FASTA format at each species page (Figure 3). At the same time, you can perform a query at the Advanced search page and download a subset of sequences in FASTA format. Moreover, everybody can also access programatically to transcript information and sequence via the REST GreeNC API.