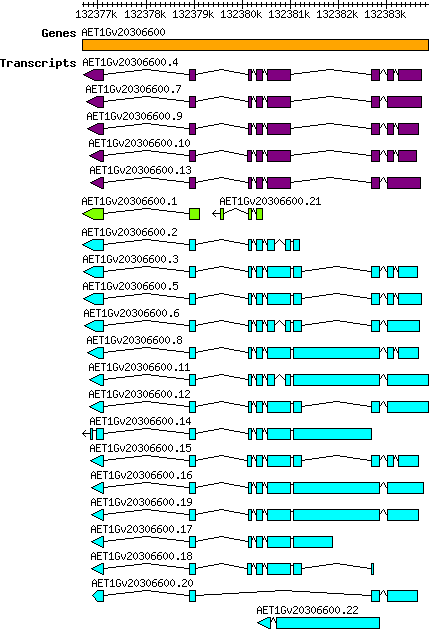
Atauschii AET1Gv20306600
From GreeNC
Gene information
| Gene alias / name | Chrom. | Start | End | Assembly | Species | Details
|
|---|---|---|---|---|---|---|
| Atauschii_AET1Gv20306600 [ AET1Gv20306600] |
1D | 132376689 | 132383860 | v4 | Aegilops tauschii (Ensembl Plants 51) | Coding? Yes |
Transcript features
This table shows how a lncRNA has been classified (high/low confidence) or whether it has been classified as pre-miRNA (go to FAQ for classification criteria). Besides, it displays the lncRNA length and sequence as well as information about the ORF, the coding potential calculated by the Coding Potential Calculator (CPC), and the folding energies calculated by RNAfold.
| Transcript alias / name | Confidence | Pre-miRNA | Length | Orthologous group | Sequence | Details
|
|---|---|---|---|---|---|---|
| Atauschii_AET1Gv20306600.1 AET1Gv20306600.1 |
High | No | 627 | OG0000758 | Get ORF Coding potential... Type: noncoding Potential: -1.089 Folding energies... MFEI: -23.493 AMFE: -0.604 GC content %: 38.915 | |
| Atauschii_AET1Gv20306600.11 AET1Gv20306600.11 |
Low | Yes | 3465 | OG0000758 | Get ORF Coding potential... Type: noncoding Potential: -1.031 Folding energies... MFEI: -32.095 AMFE: -0.687 GC content %: 46.696 | |
| Atauschii_AET1Gv20306600.12 AET1Gv20306600.12 |
Low | Yes | 2240 | OG0000758 | Get ORF Coding potential... Type: noncoding Potential: -0.995 Folding energies... MFEI: -36.893 AMFE: -0.697 GC content %: 52.946 | |
| Atauschii_AET1Gv20306600.14 AET1Gv20306600.14 |
Low | Yes | 2550 | OG0000758 | Get ORF Coding potential... Type: noncoding Potential: -1.226 Folding energies... MFEI: -29.094 AMFE: -0.718 GC content %: 40.549 | |
| Atauschii_AET1Gv20306600.15 AET1Gv20306600.15 |
Low | Yes | 1905 | OG0000758 | Get ORF Coding potential... Type: noncoding Potential: -1.201 Folding energies... MFEI: -36.147 AMFE: -0.696 GC content %: 51.916 | |
| Atauschii_AET1Gv20306600.16 AET1Gv20306600.16 |
Low | Yes | 3561 | OG0000758 | Get ORF Coding potential... Type: noncoding Potential: -1.033 Folding energies... MFEI: -32.008 AMFE: -0.695 GC content %: 46.083 | |
| Atauschii_AET1Gv20306600.17 AET1Gv20306600.17 |
Low | Yes | 1833 | OG0000758 | Get ORF Coding potential... Type: noncoding Potential: -1.203 Folding energies... MFEI: -31.037 AMFE: -0.735 GC content %: 42.226 | |
| Atauschii_AET1Gv20306600.18 AET1Gv20306600.18 |
Low | Yes | 1221 | OG0000758 | Get ORF Coding potential... Type: noncoding Potential: -1.198 Folding energies... MFEI: -33.767 AMFE: -0.739 GC content %: 45.700 | |
| Atauschii_AET1Gv20306600.19 AET1Gv20306600.19 |
Low | Yes | 3438 | OG0000758 | Get ORF Coding potential... Type: noncoding Potential: -1.241 Folding energies... MFEI: -31.024 AMFE: -0.683 GC content %: 45.404 | |
| Atauschii_AET1Gv20306600.2 AET1Gv20306600.2 |
Low | Yes | 1100 | OG0000758 | Get ORF Coding potential... Type: noncoding Potential: -1.086 Folding energies... MFEI: -32.364 AMFE: -0.713 GC content %: 45.364 | |
| Atauschii_AET1Gv20306600.20 AET1Gv20306600.20 |
Low | No | 1111 | OG0000758 | Get ORF Coding potential... Type: noncoding Potential: -1.071 Folding energies... MFEI: -33.780 AMFE: -0.612 GC content %: 55.176 | |
| Atauschii_AET1Gv20306600.21 AET1Gv20306600.21 |
High | No | 252 | OG0068138 | Get ORF Coding potential... Type: noncoding Potential: -1.053 Folding energies... MFEI: -23.929 AMFE: -0.543 GC content %: 44.048 | |
| Atauschii_AET1Gv20306600.22 AET1Gv20306600.22 |
Low | No | 2422 | OG0000758 | Get ORF Coding potential... Type: noncoding Potential: -0.882 Folding energies... MFEI: -26.090 AMFE: -0.633 GC content %: 41.206 | |
| Atauschii_AET1Gv20306600.3 AET1Gv20306600.3 |
Low | Yes | 2024 | OG0000758 | Get ORF Coding potential... Type: noncoding Potential: -1.210 Folding energies... MFEI: -35.124 AMFE: -0.693 GC content %: 50.692 | |
| Atauschii_AET1Gv20306600.5 AET1Gv20306600.5 |
Low | Yes | 2102 | OG0000758 | Get ORF Coding potential... Type: noncoding Potential: -1.025 Folding energies... MFEI: -36.032 AMFE: -0.700 GC content %: 51.475 | |
| Atauschii_AET1Gv20306600.6 AET1Gv20306600.6 |
Low | Yes | 1917 | OG0000758 | Get ORF Coding potential... Type: noncoding Potential: -1.202 Folding energies... MFEI: -35.347 AMFE: -0.671 GC content %: 52.686 | |
| Atauschii_AET1Gv20306600.8 AET1Gv20306600.8 |
Low | Yes | 3416 | OG0000758 | Get ORF Coding potential... Type: noncoding Potential: -1.241 Folding energies... MFEI: -31.156 AMFE: -0.693 GC content %: 44.936 |
Matches to external databases
| Transcript alias | Database | Version | Hit | Evalue |
|---|---|---|---|---|
| Atauschii_AET1Gv20306600.15 | SwissProt | 2021-04-01 | Q2PS26 | 6.96e-5 |
| Atauschii_AET1Gv20306600.17 | Rfam | 14.6 | mir-1253 | 1.3e-10 |
| Atauschii_AET1Gv20306600.11 | Rfam | 14.6 | mir-1253 | 2.5e-10 |
| Atauschii_AET1Gv20306600.22 | SwissProt | 2021-04-01 | Q7XR61 | 5.7e-8 |
| Atauschii_AET1Gv20306600.6 | Rfam | 14.6 | mir-1253 | 1.4e-10 |
| Atauschii_AET1Gv20306600.16 | SwissProt | 2021-04-01 | Q7XR61 | 9.37e-8 |
| Atauschii_AET1Gv20306600.18 | Rfam | 14.6 | mir-1253 | 8.8e-11 |
| Atauschii_AET1Gv20306600.12 | Rfam | 14.6 | mir-1253 | 1.6e-10 |
| Atauschii_AET1Gv20306600.3 | SwissProt | 2021-04-01 | Q2PS26 | 7.56e-5 |
| Atauschii_AET1Gv20306600.8 | Rfam | 14.6 | mir-1253 | 2.5e-10 |
... further results
Gene models