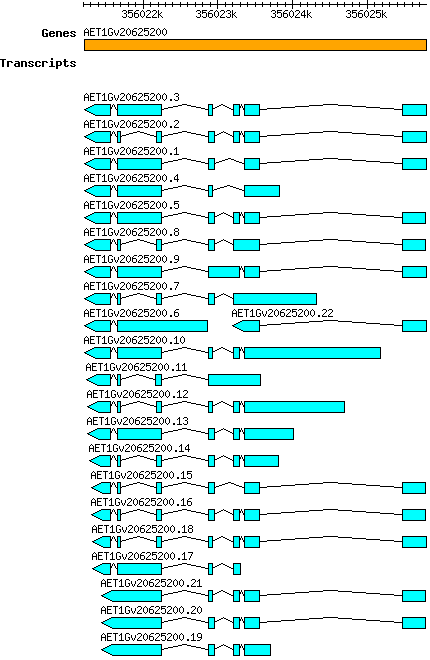
Atauschii AET1Gv20625200
From GreeNC
Gene information
| Gene alias / name | Chrom. | Start | End | Assembly | Species | Details
|
|---|---|---|---|---|---|---|
| Atauschii_AET1Gv20625200 [ AET1Gv20625200] |
1D | 356021213 | 356025794 | v4 | Aegilops tauschii (Ensembl Plants 51) | Coding? No |
Transcript features
This table shows how a lncRNA has been classified (high/low confidence) or whether it has been classified as pre-miRNA (go to FAQ for classification criteria). Besides, it displays the lncRNA length and sequence as well as information about the ORF, the coding potential calculated by the Coding Potential Calculator (CPC), and the folding energies calculated by RNAfold.
| Transcript alias / name | Confidence | Pre-miRNA | Length | Orthologous group | Sequence | Details
|
|---|---|---|---|---|---|---|
| Atauschii_AET1Gv20625200.1 AET1Gv20625200.1 |
Low | No | 1533 | OG0000392 | Get ORF Coding potential... Type: noncoding Potential: -1.112 Folding energies... MFEI: -28.023 AMFE: -0.661 GC content %: 42.401 | |
| Atauschii_AET1Gv20625200.10 AET1Gv20625200.10 |
Low | No | 2850 | OG0000392 | Get ORF Coding potential... Type: noncoding Potential: -1.204 Folding energies... MFEI: -26.439 AMFE: -0.673 GC content %: 39.263 | |
| Atauschii_AET1Gv20625200.11 AET1Gv20625200.11 |
Low | No | 1114 | OG0000392 | Get ORF Coding potential... Type: noncoding Potential: -1.205 Folding energies... MFEI: -23.986 AMFE: -0.611 GC content %: 39.228 | |
| Atauschii_AET1Gv20625200.12 AET1Gv20625200.12 |
Low | No | 1858 | OG0000392 | Get ORF Coding potential... Type: noncoding Potential: -1.189 Folding energies... MFEI: -24.672 AMFE: -0.647 GC content %: 38.105 | |
| Atauschii_AET1Gv20625200.13 AET1Gv20625200.13 |
Low | No | 1693 | OG0000392 | Get ORF Coding potential... Type: noncoding Potential: -1.130 Folding energies... MFEI: -24.962 AMFE: -0.655 GC content %: 38.098 | |
| Atauschii_AET1Gv20625200.14 AET1Gv20625200.14 |
Low | No | 985 | OG0000392 | Get ORF Coding potential... Type: noncoding Potential: -1.063 Folding energies... MFEI: -24.091 AMFE: -0.602 GC content %: 40.000 | |
| Atauschii_AET1Gv20625200.15 AET1Gv20625200.15 |
Low | No | 945 | OG0000392 | Get ORF Coding potential... Type: noncoding Potential: -1.179 Folding energies... MFEI: -28.392 AMFE: -0.611 GC content %: 46.455 | |
| Atauschii_AET1Gv20625200.16 AET1Gv20625200.16 |
Low | No | 1018 | OG0000392 | Get ORF Coding potential... Type: noncoding Potential: -0.955 Folding energies... MFEI: -28.487 AMFE: -0.613 GC content %: 46.464 | |
| Atauschii_AET1Gv20625200.17 AET1Gv20625200.17 |
Low | No | 955 | OG0000392 | Get ORF Coding potential... Type: noncoding Potential: -1.007 Folding energies... MFEI: -23.958 AMFE: -0.652 GC content %: 36.754 | |
| Atauschii_AET1Gv20625200.18 AET1Gv20625200.18 |
Low | No | 995 | OG0000392 | Get ORF Coding potential... Type: noncoding Potential: -1.107 Folding energies... MFEI: -29.357 AMFE: -0.621 GC content %: 47.236 | |
| Atauschii_AET1Gv20625200.19 AET1Gv20625200.19 |
Low | No | 1299 | OG0000392 | Get ORF Coding potential... Type: noncoding Potential: -1.079 Folding energies... MFEI: -24.650 AMFE: -0.634 GC content %: 38.876 | |
| Atauschii_AET1Gv20625200.2 AET1Gv20625200.2 |
Low | No | 1131 | OG0000392 | Get ORF Coding potential... Type: noncoding Potential: -1.132 Folding energies... MFEI: -29.089 AMFE: -0.630 GC content %: 46.154 | |
| Atauschii_AET1Gv20625200.20 AET1Gv20625200.20 |
Low | No | 1462 | OG0000392 | Get ORF Coding potential... Type: noncoding Potential: -1.103 Folding energies... MFEI: -27.517 AMFE: -0.640 GC content %: 43.023 | |
| Atauschii_AET1Gv20625200.21 AET1Gv20625200.21 |
Low | No | 1429 | OG0000392 | Get ORF Coding potential... Type: noncoding Potential: -1.098 Folding energies... MFEI: -27.782 AMFE: -0.646 GC content %: 43.037 | |
| Atauschii_AET1Gv20625200.22 AET1Gv20625200.22 |
Low | No | 697 | OG0000392 | Get ORF Coding potential... Type: noncoding Potential: -1.090 Folding energies... MFEI: -31.521 AMFE: -0.624 GC content %: 50.502 | |
| Atauschii_AET1Gv20625200.3 AET1Gv20625200.3 |
Low | No | 1572 | OG0000392 | Get ORF Coding potential... Type: noncoding Potential: -1.116 Folding energies... MFEI: -28.658 AMFE: -0.673 GC content %: 42.557 | |
| Atauschii_AET1Gv20625200.4 AET1Gv20625200.4 |
Low | No | 1447 | OG0000392 | Get ORF Coding potential... Type: noncoding Potential: -1.101 Folding energies... MFEI: -25.197 AMFE: -0.667 GC content %: 37.802 | |
| Atauschii_AET1Gv20625200.5 AET1Gv20625200.5 |
Low | No | 1596 | OG0000392 | Get ORF Coding potential... Type: noncoding Potential: -1.119 Folding energies... MFEI: -28.503 AMFE: -0.673 GC content %: 42.356 | |
| Atauschii_AET1Gv20625200.6 AET1Gv20625200.6 |
Low | No | 1552 | OG0000392 | Get ORF Coding potential... Type: noncoding Potential: -1.114 Folding energies... MFEI: -22.726 AMFE: -0.654 GC content %: 34.729 | |
| Atauschii_AET1Gv20625200.7 AET1Gv20625200.7 |
Low | No | 1635 | OG0000392 | Get ORF Coding potential... Type: noncoding Potential: -1.222 Folding energies... MFEI: -24.110 AMFE: -0.623 GC content %: 38.716 | |
| Atauschii_AET1Gv20625200.8 AET1Gv20625200.8 |
Low | No | 1191 | OG0000392 | Get ORF Coding potential... Type: noncoding Potential: -1.200 Folding energies... MFEI: -28.296 AMFE: -0.629 GC content %: 45.004 | |
| Atauschii_AET1Gv20625200.9 AET1Gv20625200.9 |
Low | No | 1859 | OG0000392 | Get ORF Coding potential... Type: noncoding Potential: -1.145 Folding energies... MFEI: -27.133 AMFE: -0.653 GC content %: 41.528 |
Matches to external databases
| Transcript alias | Database | Version | Hit | Evalue |
|---|---|---|---|---|
| Atauschii_AET1Gv20625200.21 | RepBase (RepeatMasker) | 2014-01-31 | DNA/CMC-EnSpm | |
| Atauschii_AET1Gv20625200.17 | RepBase (RepeatMasker) | 2014-01-31 | DNA/CMC-EnSpm | |
| Atauschii_AET1Gv20625200.12 | RepBase (RepeatMasker) | 2014-01-31 | DNA/CMC-EnSpm | |
| Atauschii_AET1Gv20625200.7 | RepBase (RepeatMasker) | 2014-01-31 | DNA/CMC-EnSpm | |
| Atauschii_AET1Gv20625200.22 | RepBase (RepeatMasker) | 2014-01-31 | DNA/CMC-EnSpm | |
| Atauschii_AET1Gv20625200.18 | RepBase (RepeatMasker) | 2014-01-31 | DNA/CMC-EnSpm | |
| Atauschii_AET1Gv20625200.13 | RepBase (RepeatMasker) | 2014-01-31 | DNA/CMC-EnSpm | |
| Atauschii_AET1Gv20625200.8 | RepBase (RepeatMasker) | 2014-01-31 | DNA/CMC-EnSpm | |
| Atauschii_AET1Gv20625200.3 | RepBase (RepeatMasker) | 2014-01-31 | DNA/CMC-EnSpm | |
| Atauschii_AET1Gv20625200.19 | RepBase (RepeatMasker) | 2014-01-31 | DNA/CMC-EnSpm |
... further results
Gene models