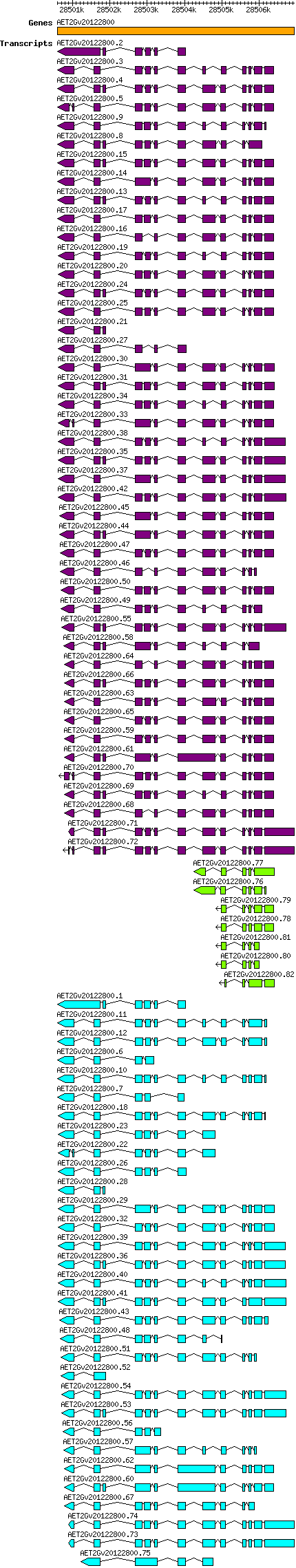
Atauschii AET2Gv20122800
From GreeNC
Gene information
| Gene alias / name | Chrom. | Start | End | Assembly | Species | Details
|
|---|---|---|---|---|---|---|
| Atauschii_AET2Gv20122800 [ AET2Gv20122800] |
2D | 28500591 | 28506938 | v4 | Aegilops tauschii (Ensembl Plants 51) | Coding? Yes |
Transcript features
This table shows how a lncRNA has been classified (high/low confidence) or whether it has been classified as pre-miRNA (go to FAQ for classification criteria). Besides, it displays the lncRNA length and sequence as well as information about the ORF, the coding potential calculated by the Coding Potential Calculator (CPC), and the folding energies calculated by RNAfold.
| Transcript alias / name | Confidence | Pre-miRNA | Length | Orthologous group | Sequence | Details
|
|---|---|---|---|---|---|---|
| Atauschii_AET2Gv20122800.1 AET2Gv20122800.1 |
Low | No | 1834 | OG0000104 | Get ORF Coding potential... Type: noncoding Potential: 0.000 Folding energies... MFEI: -28.697 AMFE: -0.713 GC content %: 40.240 | |
| Atauschii_AET2Gv20122800.10 AET2Gv20122800.10 |
Low | No | 1823 | OG0000104 | Get ORF Coding potential... Type: noncoding Potential: -0.212 Folding energies... MFEI: -32.830 AMFE: -0.709 GC content %: 46.297 | |
| Atauschii_AET2Gv20122800.11 AET2Gv20122800.11 |
Low | No | 1862 | OG0000104 | Get ORF Coding potential... Type: noncoding Potential: -0.188 Folding energies... MFEI: -32.696 AMFE: -0.704 GC content %: 46.455 | |
| Atauschii_AET2Gv20122800.12 AET2Gv20122800.12 |
Low | No | 2149 | OG0000104 | Get ORF Coding potential... Type: noncoding Potential: -0.138 Folding energies... MFEI: -32.080 AMFE: -0.709 GC content %: 45.230 | |
| Atauschii_AET2Gv20122800.18 AET2Gv20122800.18 |
Low | No | 2033 | OG0000104 | Get ORF Coding potential... Type: noncoding Potential: -0.136 Folding energies... MFEI: -32.115 AMFE: -0.709 GC content %: 45.303 | |
| Atauschii_AET2Gv20122800.22 AET2Gv20122800.22 |
Low | No | 1418 | OG0000104 | Get ORF Coding potential... Type: noncoding Potential: -0.189 Folding energies... MFEI: -29.972 AMFE: -0.684 GC content %: 43.794 | |
| Atauschii_AET2Gv20122800.23 AET2Gv20122800.23 |
Low | No | 1539 | OG0000104 | Get ORF Coding potential... Type: noncoding Potential: -0.149 Folding energies... MFEI: -30.747 AMFE: -0.703 GC content %: 43.730 | |
| Atauschii_AET2Gv20122800.26 AET2Gv20122800.26 |
Low | No | 1213 | OG0000104 | Get ORF Coding potential... Type: noncoding Potential: -0.182 Folding energies... MFEI: -30.676 AMFE: -0.692 GC content %: 44.353 | |
| Atauschii_AET2Gv20122800.28 AET2Gv20122800.28 |
Low | No | 644 | OG0000104 | Get ORF Coding potential... Type: noncoding Potential: -0.012 Folding energies... MFEI: -27.717 AMFE: -0.720 GC content %: 38.509 | |
| Atauschii_AET2Gv20122800.29 AET2Gv20122800.29 |
Low | No | 2346 | OG0000104 | Get ORF Coding potential... Type: noncoding Potential: -0.002 Folding energies... MFEI: -33.559 AMFE: -0.711 GC content %: 47.187 | |
| Atauschii_AET2Gv20122800.32 AET2Gv20122800.32 |
Low | No | 2228 | OG0000104 | Get ORF Coding potential... Type: noncoding Potential: 0.000 Folding energies... MFEI: -33.842 AMFE: -0.710 GC content %: 47.666 | |
| Atauschii_AET2Gv20122800.36 AET2Gv20122800.36 |
Low | No | 2637 | OG0000104 | Get ORF Coding potential... Type: noncoding Potential: -0.054 Folding energies... MFEI: -33.534 AMFE: -0.698 GC content %: 48.009 | |
| Atauschii_AET2Gv20122800.39 AET2Gv20122800.39 |
Low | No | 2554 | OG0000104 | Get ORF Coding potential... Type: noncoding Potential: -0.014 Folding energies... MFEI: -34.636 AMFE: -0.715 GC content %: 48.473 | |
| Atauschii_AET2Gv20122800.40 AET2Gv20122800.40 |
Low | No | 2277 | OG0000104 | Get ORF Coding potential... Type: noncoding Potential: -0.001 Folding energies... MFEI: -35.152 AMFE: -0.706 GC content %: 49.802 | |
| Atauschii_AET2Gv20122800.41 AET2Gv20122800.41 |
Low | No | 2737 | OG0000104 | Get ORF Coding potential... Type: noncoding Potential: -0.019 Folding energies... MFEI: -33.822 AMFE: -0.707 GC content %: 47.863 | |
| Atauschii_AET2Gv20122800.43 AET2Gv20122800.43 |
Low | No | 2050 | OG0000104 | Get ORF Coding potential... Type: noncoding Potential: -0.168 Folding energies... MFEI: -32.351 AMFE: -0.700 GC content %: 46.195 | |
| Atauschii_AET2Gv20122800.48 AET2Gv20122800.48 |
Low | No | 1233 | OG0000104 | Get ORF Coding potential... Type: noncoding Potential: -0.190 Folding energies... MFEI: -31.817 AMFE: -0.701 GC content %: 45.418 | |
| Atauschii_AET2Gv20122800.51 AET2Gv20122800.51 |
Low | No | 1763 | OG0000104 | Get ORF Coding potential... Type: noncoding Potential: -0.227 Folding energies... MFEI: -30.647 AMFE: -0.694 GC content %: 44.186 | |
| Atauschii_AET2Gv20122800.52 AET2Gv20122800.52 |
Low | No | 658 | OG0000104 | Get ORF Coding potential... Type: noncoding Potential: -0.078 Folding energies... MFEI: -28.784 AMFE: -0.709 GC content %: 40.578 | |
| Atauschii_AET2Gv20122800.53 AET2Gv20122800.53 |
Low | No | 2556 | OG0000104 | Get ORF Coding potential... Type: noncoding Potential: -0.048 Folding energies... MFEI: -33.791 AMFE: -0.694 GC content %: 48.709 | |
| Atauschii_AET2Gv20122800.54 AET2Gv20122800.54 |
Low | No | 2448 | OG0000104 | Get ORF Coding potential... Type: noncoding Potential: -0.009 Folding energies... MFEI: -34.285 AMFE: -0.697 GC content %: 49.224 | |
| Atauschii_AET2Gv20122800.56 AET2Gv20122800.56 |
Low | No | 962 | OG0000104 | Get ORF Coding potential... Type: noncoding Potential: -0.217 Folding energies... MFEI: -30.343 AMFE: -0.674 GC content %: 45.010 | |
| Atauschii_AET2Gv20122800.57 AET2Gv20122800.57 |
Low | No | 1514 | OG0000104 | Get ORF Coding potential... Type: noncoding Potential: -0.059 Folding energies... MFEI: -31.757 AMFE: -0.696 GC content %: 45.641 | |
| Atauschii_AET2Gv20122800.6 AET2Gv20122800.6 |
Low | No | 1006 | OG0000104 | Get ORF Coding potential... Type: noncoding Potential: -0.217 Folding energies... MFEI: -30.954 AMFE: -0.709 GC content %: 43.638 | |
| Atauschii_AET2Gv20122800.60 AET2Gv20122800.60 |
Low | No | 2704 | OG0000104 | Get ORF Coding potential... Type: noncoding Potential: -0.011 Folding energies... MFEI: -32.518 AMFE: -0.708 GC content %: 45.932 | |
| Atauschii_AET2Gv20122800.62 AET2Gv20122800.62 |
Low | No | 2654 | OG0000104 | Get ORF Coding potential... Type: noncoding Potential: -0.016 Folding energies... MFEI: -32.513 AMFE: -0.705 GC content %: 46.119 | |
| Atauschii_AET2Gv20122800.67 AET2Gv20122800.67 |
Low | No | 1703 | OG0000104 | Get ORF Coding potential... Type: noncoding Potential: -0.241 Folding energies... MFEI: -31.409 AMFE: -0.713 GC content %: 44.040 | |
| Atauschii_AET2Gv20122800.7 AET2Gv20122800.7 |
Low | No | 1089 | OG0000104 | Get ORF Coding potential... Type: noncoding Potential: -0.254 Folding energies... MFEI: -30.918 AMFE: -0.697 GC content %: 44.353 | |
| Atauschii_AET2Gv20122800.73 AET2Gv20122800.73 |
Low | No | 2537 | OG0000104 | Get ORF Coding potential... Type: noncoding Potential: -0.014 Folding energies... MFEI: -34.754 AMFE: -0.689 GC content %: 50.453 | |
| Atauschii_AET2Gv20122800.74 AET2Gv20122800.74 |
Low | No | 2541 | OG0000104 | Get ORF Coding potential... Type: noncoding Potential: -0.014 Folding energies... MFEI: -34.864 AMFE: -0.691 GC content %: 50.453 | |
| Atauschii_AET2Gv20122800.75 AET2Gv20122800.75 |
Low | No | 1573 | OG0000104 | Get ORF Coding potential... Type: noncoding Potential: -0.239 Folding energies... MFEI: -30.337 AMFE: -0.704 GC content %: 43.102 | |
| Atauschii_AET2Gv20122800.76 AET2Gv20122800.76 |
High | No | 1110 | OG0000104 | Get ORF Coding potential... Type: noncoding Potential: -0.831 Folding energies... MFEI: -29.766 AMFE: -0.672 GC content %: 44.324 | |
| Atauschii_AET2Gv20122800.77 AET2Gv20122800.77 |
High | No | 1137 | OG0000104 | Get ORF Coding potential... Type: noncoding Potential: -0.790 Folding energies... MFEI: -34.037 AMFE: -0.681 GC content %: 49.956 | |
| Atauschii_AET2Gv20122800.78 AET2Gv20122800.78 |
High | No | 740 | OG0000104 | Get ORF Coding potential... Type: noncoding Potential: -0.908 Folding energies... MFEI: -37.811 AMFE: -0.682 GC content %: 55.405 | |
| Atauschii_AET2Gv20122800.79 AET2Gv20122800.79 |
High | No | 709 | OG0000104 | Get ORF Coding potential... Type: noncoding Potential: -0.733 Folding energies... MFEI: -38.815 AMFE: -0.691 GC content %: 56.135 | |
| Atauschii_AET2Gv20122800.80 AET2Gv20122800.80 |
High | No | 401 | OG0024538 | Get ORF Coding potential... Type: noncoding Potential: -0.702 Folding energies... MFEI: -26.758 AMFE: -0.571 GC content %: 46.883 | |
| Atauschii_AET2Gv20122800.81 AET2Gv20122800.81 |
High | No | 374 | OG0024538 | Get ORF Coding potential... Type: noncoding Potential: -0.748 Folding energies... MFEI: -27.701 AMFE: -0.582 GC content %: 47.594 | |
| Atauschii_AET2Gv20122800.82 AET2Gv20122800.82 |
High | No | 702 | OG0000104 | Get ORF Coding potential... Type: noncoding Potential: -0.903 Folding energies... MFEI: -39.843 AMFE: -0.696 GC content %: 57.265 |
Matches to external databases
| Transcript alias | Database | Version | Hit | Evalue |
|---|---|---|---|---|
| Atauschii_AET2Gv20122800.39 | SwissProt | 2021-04-01 | O04658 | 1.07e-17 |
| Atauschii_AET2Gv20122800.73 | SwissProt | 2021-04-01 | O04658 | 1.05e-17 |
| Atauschii_AET2Gv20122800.26 | SwissProt | 2021-04-01 | O04658 | 1.45e-18 |
| Atauschii_AET2Gv20122800.6 | SwissProt | 2021-04-01 | O04658 | 5.29e-19 |
| Atauschii_AET2Gv20122800.11 | SwissProt | 2021-04-01 | O04658 | 5.34e-18 |
| Atauschii_AET2Gv20122800.52 | SwissProt | 2021-04-01 | O04658 | 1.53e-25 |
| Atauschii_AET2Gv20122800.40 | SwissProt | 2021-04-01 | O04658 | 7.99e-18 |
| Atauschii_AET2Gv20122800.74 | SwissProt | 2021-04-01 | O04658 | 1.07e-17 |
| Atauschii_AET2Gv20122800.28 | SwissProt | 2021-04-01 | O04658 | 6.31e-29 |
| Atauschii_AET2Gv20122800.60 | SwissProt | 2021-04-01 | O04658 | 1.32e-27 |
... further results
Gene models