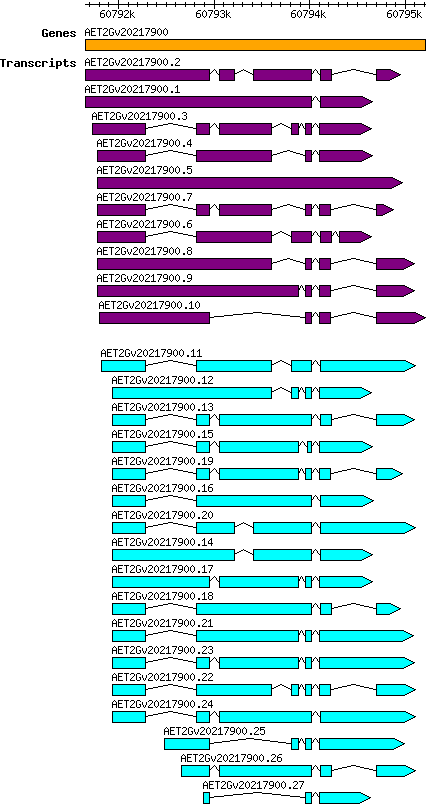
Atauschii AET2Gv20217900
From GreeNC
Gene information
| Gene alias / name | Chrom. | Start | End | Assembly | Species | Details
|
|---|---|---|---|---|---|---|
| Atauschii_AET2Gv20217900 [ AET2Gv20217900] |
2D | 60791653 | 60795214 | v4 | Aegilops tauschii (Ensembl Plants 51) | Coding? Yes |
Transcript features
This table shows how a lncRNA has been classified (high/low confidence) or whether it has been classified as pre-miRNA (go to FAQ for classification criteria). Besides, it displays the lncRNA length and sequence as well as information about the ORF, the coding potential calculated by the Coding Potential Calculator (CPC), and the folding energies calculated by RNAfold.
| Transcript alias / name | Confidence | Pre-miRNA | Length | Orthologous group | Sequence | Details
|
|---|---|---|---|---|---|---|
| Atauschii_AET2Gv20217900.11 AET2Gv20217900.11 |
Low | No | 2446 | OG0000682 | Get ORF Coding potential... Type: noncoding Potential: -1.230 Folding energies... MFEI: -28.373 AMFE: -0.602 GC content %: 47.138 | |
| Atauschii_AET2Gv20217900.12 AET2Gv20217900.12 |
Low | No | 2340 | OG0000682 | Get ORF Coding potential... Type: noncoding Potential: -1.142 Folding energies... MFEI: -28.915 AMFE: -0.637 GC content %: 45.427 | |
| Atauschii_AET2Gv20217900.13 AET2Gv20217900.13 |
Low | No | 1963 | OG0000682 | Get ORF Coding potential... Type: noncoding Potential: -1.140 Folding energies... MFEI: -28.207 AMFE: -0.626 GC content %: 45.084 | |
| Atauschii_AET2Gv20217900.14 AET2Gv20217900.14 |
Low | No | 2431 | OG0000682 | Get ORF Coding potential... Type: noncoding Potential: -1.147 Folding energies... MFEI: -28.626 AMFE: -0.639 GC content %: 44.796 | |
| Atauschii_AET2Gv20217900.15 AET2Gv20217900.15 |
Low | No | 1897 | OG0000682 | Get ORF Coding potential... Type: noncoding Potential: -1.047 Folding energies... MFEI: -29.025 AMFE: -0.617 GC content %: 47.022 | |
| Atauschii_AET2Gv20217900.16 AET2Gv20217900.16 |
Low | No | 2102 | OG0000682 | Get ORF Coding potential... Type: noncoding Potential: -1.222 Folding energies... MFEI: -28.302 AMFE: -0.618 GC content %: 45.814 | |
| Atauschii_AET2Gv20217900.17 AET2Gv20217900.17 |
Low | No | 2452 | OG0000682 | Get ORF Coding potential... Type: noncoding Potential: -1.148 Folding energies... MFEI: -28.723 AMFE: -0.644 GC content %: 44.617 | |
| Atauschii_AET2Gv20217900.18 AET2Gv20217900.18 |
Low | No | 1915 | OG0000682 | Get ORF Coding potential... Type: noncoding Potential: -1.139 Folding energies... MFEI: -28.339 AMFE: -0.625 GC content %: 45.326 | |
| Atauschii_AET2Gv20217900.19 AET2Gv20217900.19 |
Low | No | 1766 | OG0000682 | Get ORF Coding potential... Type: noncoding Potential: -1.135 Folding energies... MFEI: -28.584 AMFE: -0.621 GC content %: 46.036 | |
| Atauschii_AET2Gv20217900.20 AET2Gv20217900.20 |
Low | No | 2352 | OG0000682 | Get ORF Coding potential... Type: noncoding Potential: -1.228 Folding energies... MFEI: -28.452 AMFE: -0.627 GC content %: 45.408 | |
| Atauschii_AET2Gv20217900.21 AET2Gv20217900.21 |
Low | No | 2459 | OG0000682 | Get ORF Coding potential... Type: noncoding Potential: -1.230 Folding energies... MFEI: -28.203 AMFE: -0.628 GC content %: 44.937 | |
| Atauschii_AET2Gv20217900.22 AET2Gv20217900.22 |
Low | No | 1779 | OG0000682 | Get ORF Coding potential... Type: noncoding Potential: -1.135 Folding energies... MFEI: -28.522 AMFE: -0.613 GC content %: 46.543 | |
| Atauschii_AET2Gv20217900.23 AET2Gv20217900.23 |
Low | No | 2361 | OG0000682 | Get ORF Coding potential... Type: noncoding Potential: -1.228 Folding energies... MFEI: -28.403 AMFE: -0.626 GC content %: 45.362 | |
| Atauschii_AET2Gv20217900.24 AET2Gv20217900.24 |
Low | No | 2436 | OG0000682 | Get ORF Coding potential... Type: noncoding Potential: -1.230 Folding energies... MFEI: -28.296 AMFE: -0.628 GC content %: 45.074 | |
| Atauschii_AET2Gv20217900.25 AET2Gv20217900.25 |
Low | No | 1497 | OG0000682 | Get ORF Coding potential... Type: noncoding Potential: -1.199 Folding energies... MFEI: -28.697 AMFE: -0.695 GC content %: 41.283 | |
| Atauschii_AET2Gv20217900.26 AET2Gv20217900.26 |
Low | No | 1782 | OG0000682 | Get ORF Coding potential... Type: noncoding Potential: -1.056 Folding energies... MFEI: -27.806 AMFE: -0.701 GC content %: 39.675 | |
| Atauschii_AET2Gv20217900.27 AET2Gv20217900.27 |
Low | No | 658 | OG0000682 | Get ORF Coding potential... Type: noncoding Potential: -0.411 Folding energies... MFEI: -28.313 AMFE: -0.642 GC content %: 44.073 |
Matches to external databases
| Transcript alias | Database | Version | Hit | Evalue |
|---|---|---|---|---|
| Atauschii_AET2Gv20217900.13 | SwissProt | 2021-04-01 | Q5NBJ3 | 4.28e-9 |
| Atauschii_AET2Gv20217900.14 | RepBase (RepeatMasker) | 2014-01-31 | DNA/CMC-Transib | |
| Atauschii_AET2Gv20217900.24 | SwissProt | 2021-04-01 | Q5NBJ3 | 3.05e-9 |
| Atauschii_AET2Gv20217900.19 | SwissProt | 2021-04-01 | Q5NBJ3 | 3.58e-9 |
| Atauschii_AET2Gv20217900.14 | SwissProt | 2021-04-01 | Q5NBJ3 | 3.04e-9 |
| Atauschii_AET2Gv20217900.17 | RepBase (RepeatMasker) | 2014-01-31 | DNA/CMC-Transib | |
| Atauschii_AET2Gv20217900.25 | SwissProt | 2021-04-01 | Q5NBJ3 | 1.33e-9 |
| Atauschii_AET2Gv20217900.20 | SwissProt | 2021-04-01 | Q5NBJ3 | 2.89e-9 |
| Atauschii_AET2Gv20217900.15 | SwissProt | 2021-04-01 | Q5NBJ3 | 8.0e-10 |
| Atauschii_AET2Gv20217900.26 | SwissProt | 2021-04-01 | Q5NBJ3 | 3.64e-9 |
... further results
Gene models