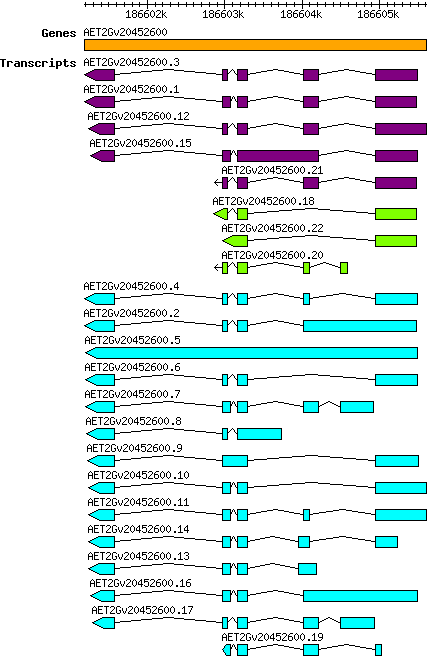
Atauschii AET2Gv20452600
From GreeNC
Gene information
| Gene alias / name | Chrom. | Start | End | Assembly | Species | Details
|
|---|---|---|---|---|---|---|
| Atauschii_AET2Gv20452600 [ AET2Gv20452600] |
2D | 186601183 | 186605609 | v4 | Aegilops tauschii (Ensembl Plants 51) | Coding? Yes |
Transcript features
This table shows how a lncRNA has been classified (high/low confidence) or whether it has been classified as pre-miRNA (go to FAQ for classification criteria). Besides, it displays the lncRNA length and sequence as well as information about the ORF, the coding potential calculated by the Coding Potential Calculator (CPC), and the folding energies calculated by RNAfold.
| Transcript alias / name | Confidence | Pre-miRNA | Length | Orthologous group | Sequence | Details
|
|---|---|---|---|---|---|---|
| Atauschii_AET2Gv20452600.10 AET2Gv20452600.10 |
Low | Yes | 1246 | OG0000987 | Get ORF Coding potential... Type: noncoding Potential: -1.137 Folding energies... MFEI: -25.457 AMFE: -0.479 GC content %: 53.130 | |
| Atauschii_AET2Gv20452600.11 AET2Gv20452600.11 |
Low | Yes | 1322 | OG0000987 | Get ORF Coding potential... Type: noncoding Potential: -1.141 Folding energies... MFEI: -25.204 AMFE: -0.484 GC content %: 52.118 | |
| Atauschii_AET2Gv20452600.13 AET2Gv20452600.13 |
Low | Yes | 801 | OG0000987 | Get ORF Coding potential... Type: noncoding Potential: -0.968 Folding energies... MFEI: -24.894 AMFE: -0.652 GC content %: 38.202 | |
| Atauschii_AET2Gv20452600.14 AET2Gv20452600.14 |
Low | Yes | 999 | OG0000987 | Get ORF Coding potential... Type: noncoding Potential: -0.995 Folding energies... MFEI: -24.184 AMFE: -0.522 GC content %: 46.346 | |
| Atauschii_AET2Gv20452600.16 AET2Gv20452600.16 |
Low | Yes | 2028 | OG0000987 | Get ORF Coding potential... Type: noncoding Potential: -1.097 Folding energies... MFEI: -24.808 AMFE: -0.552 GC content %: 44.921 | |
| Atauschii_AET2Gv20452600.17 AET2Gv20452600.17 |
Low | Yes | 1118 | OG0000987 | Get ORF Coding potential... Type: noncoding Potential: -1.057 Folding energies... MFEI: -26.351 AMFE: -0.705 GC content %: 37.388 | |
| Atauschii_AET2Gv20452600.18 AET2Gv20452600.18 |
High | No | 838 | OG0070313 | Get ORF Coding potential... Type: noncoding Potential: -1.140 Folding energies... MFEI: -22.053 AMFE: -0.387 GC content %: 56.921 | |
| Atauschii_AET2Gv20452600.19 AET2Gv20452600.19 |
Low | No | 522 | OG0070314 | Get ORF Coding potential... Type: noncoding Potential: -0.918 Folding energies... MFEI: -24.119 AMFE: -0.557 GC content %: 43.295 | |
| Atauschii_AET2Gv20452600.2 AET2Gv20452600.2 |
Low | Yes | 2055 | OG0000987 | Get ORF Coding potential... Type: noncoding Potential: -1.098 Folding energies... MFEI: -24.686 AMFE: -0.554 GC content %: 44.526 | |
| Atauschii_AET2Gv20452600.20 AET2Gv20452600.20 |
High | No | 361 | OG0070315 | Get ORF Coding potential... Type: noncoding Potential: -1.077 Folding energies... MFEI: -19.391 AMFE: -0.496 GC content %: 39.058 | |
| Atauschii_AET2Gv20452600.22 AET2Gv20452600.22 |
High | No | 863 | OG0000987 | Get ORF Coding potential... Type: noncoding Potential: -1.148 Folding energies... MFEI: -21.054 AMFE: -0.371 GC content %: 56.779 | |
| Atauschii_AET2Gv20452600.4 AET2Gv20452600.4 |
Low | Yes | 1210 | OG0000987 | Get ORF Coding potential... Type: noncoding Potential: -1.231 Folding energies... MFEI: -24.463 AMFE: -0.476 GC content %: 51.405 | |
| Atauschii_AET2Gv20452600.5 AET2Gv20452600.5 |
Low | Yes | 4294 | OG0000987 | Get ORF Coding potential... Type: noncoding Potential: -1.144 Folding energies... MFEI: -25.394 AMFE: -0.629 GC content %: 40.359 | |
| Atauschii_AET2Gv20452600.6 AET2Gv20452600.6 |
Low | Yes | 1120 | OG0000987 | Get ORF Coding potential... Type: noncoding Potential: -1.214 Folding energies... MFEI: -24.616 AMFE: -0.466 GC content %: 52.857 | |
| Atauschii_AET2Gv20452600.7 AET2Gv20452600.7 |
Low | Yes | 1221 | OG0000987 | Get ORF Coding potential... Type: noncoding Potential: -1.068 Folding energies... MFEI: -26.486 AMFE: -0.708 GC content %: 37.428 | |
| Atauschii_AET2Gv20452600.8 AET2Gv20452600.8 |
Low | Yes | 1008 | OG0000987 | Get ORF Coding potential... Type: noncoding Potential: -1.188 Folding energies... MFEI: -25.714 AMFE: -0.701 GC content %: 36.706 | |
| Atauschii_AET2Gv20452600.9 AET2Gv20452600.9 |
Low | Yes | 1222 | OG0000987 | Get ORF Coding potential... Type: noncoding Potential: -1.234 Folding energies... MFEI: -23.822 AMFE: -0.463 GC content %: 51.473 |
Matches to external databases
| Transcript alias | Database | Version | Hit | Evalue |
|---|---|---|---|---|
| Atauschii_AET2Gv20452600.7 | SwissProt | 2021-04-01 | Q9SAC4 | 3.92e-5 |
| Atauschii_AET2Gv20452600.13 | RepBase (RepeatMasker) | 2014-01-31 | DNA/TcMar-Stowaway | |
| Atauschii_AET2Gv20452600.16 | SwissProt | 2021-04-01 | Q9SAC4 | 1.35e-4 |
| Atauschii_AET2Gv20452600.7 | Rfam | 14.6 | MIR1122 | 5.1e-12 |
| Atauschii_AET2Gv20452600.17 | Rfam | 14.6 | MIR1122 | 4.7e-12 |
| Atauschii_AET2Gv20452600.5 | RepBase (RepeatMasker) | 2014-01-31 | DNA/TcMar-Stowaway | |
| Atauschii_AET2Gv20452600.10 | Rfam | 14.6 | MIR1122 | 5.2e-12 |
| Atauschii_AET2Gv20452600.14 | RepBase (RepeatMasker) | 2014-01-31 | DNA/TcMar-Stowaway | |
| Atauschii_AET2Gv20452600.17 | SwissProt | 2021-04-01 | Q9SAC4 | 3.34e-5 |
| Atauschii_AET2Gv20452600.8 | Rfam | 14.6 | MIR1122 | 4.2e-12 |
... further results
Gene models