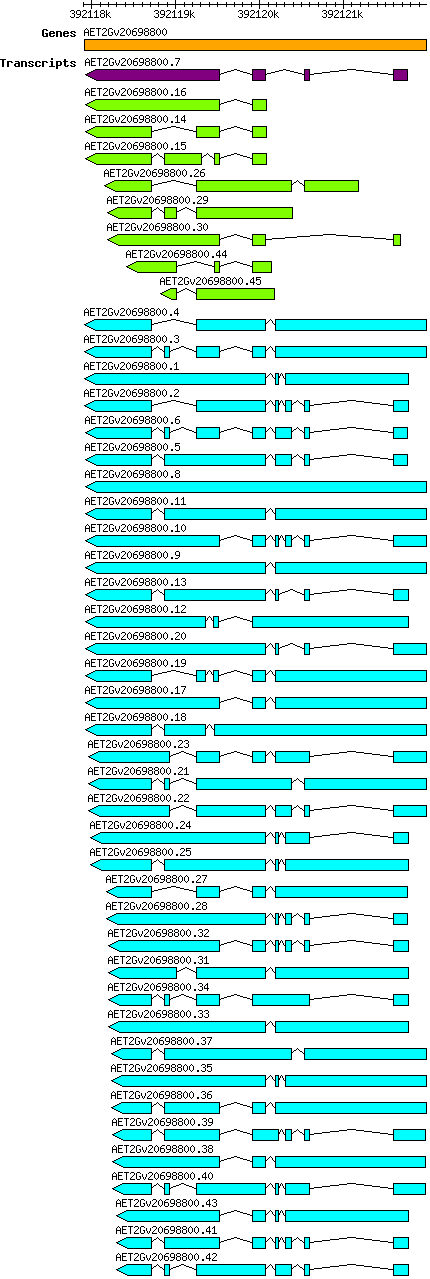
Atauschii AET2Gv20698800
From GreeNC
Gene information
| Gene alias / name | Chrom. | Start | End | Assembly | Species | Details
|
|---|---|---|---|---|---|---|
| Atauschii_AET2Gv20698800 [ AET2Gv20698800] |
2D | 392117916 | 392121985 | v4 | Aegilops tauschii (Ensembl Plants 51) | Coding? Yes |
Transcript features
This table shows how a lncRNA has been classified (high/low confidence) or whether it has been classified as pre-miRNA (go to FAQ for classification criteria). Besides, it displays the lncRNA length and sequence as well as information about the ORF, the coding potential calculated by the Coding Potential Calculator (CPC), and the folding energies calculated by RNAfold.
| Transcript alias / name | Confidence | Pre-miRNA | Length | Orthologous group | Sequence | Details
|
|---|---|---|---|---|---|---|
| Atauschii_AET2Gv20698800.1 AET2Gv20698800.1 |
Low | No | 3653 | OG0000052 | Get ORF Coding potential... Type: noncoding Potential: -1.139 Folding energies... MFEI: -28.670 AMFE: -0.686 GC content %: 41.801 | |
| Atauschii_AET2Gv20698800.10 AET2Gv20698800.10 |
Low | No | 2314 | OG0000052 | Get ORF Coding potential... Type: noncoding Potential: -1.049 Folding energies... MFEI: -30.994 AMFE: -0.665 GC content %: 46.586 | |
| Atauschii_AET2Gv20698800.11 AET2Gv20698800.11 |
Low | No | 3776 | OG0000052 | Get ORF Coding potential... Type: noncoding Potential: -1.005 Folding energies... MFEI: -29.902 AMFE: -0.687 GC content %: 43.512 | |
| Atauschii_AET2Gv20698800.12 AET2Gv20698800.12 |
Low | No | 3341 | OG0000052 | Get ORF Coding potential... Type: noncoding Potential: -1.133 Folding energies... MFEI: -28.764 AMFE: -0.683 GC content %: 42.143 | |
| Atauschii_AET2Gv20698800.13 AET2Gv20698800.13 |
Low | No | 2250 | OG0000052 | Get ORF Coding potential... Type: noncoding Potential: -1.139 Folding energies... MFEI: -29.942 AMFE: -0.683 GC content %: 43.867 | |
| Atauschii_AET2Gv20698800.14 AET2Gv20698800.14 |
High | No | 1210 | OG0000052 | Get ORF Coding potential... Type: noncoding Potential: -1.290 Folding energies... MFEI: -30.182 AMFE: -0.668 GC content %: 45.207 | |
| Atauschii_AET2Gv20698800.15 AET2Gv20698800.15 |
High | No | 1448 | OG0000052 | Get ORF Coding potential... Type: noncoding Potential: -1.308 Folding energies... MFEI: -27.914 AMFE: -0.644 GC content %: 43.370 | |
| Atauschii_AET2Gv20698800.16 AET2Gv20698800.16 |
High | No | 1756 | OG0000052 | Get ORF Coding potential... Type: noncoding Potential: -1.324 Folding energies... MFEI: -27.511 AMFE: -0.654 GC content %: 42.084 | |
| Atauschii_AET2Gv20698800.17 AET2Gv20698800.17 |
Low | No | 3536 | OG0000052 | Get ORF Coding potential... Type: noncoding Potential: -0.994 Folding energies... MFEI: -29.579 AMFE: -0.678 GC content %: 43.609 | |
| Atauschii_AET2Gv20698800.18 AET2Gv20698800.18 |
Low | No | 3788 | OG0000052 | Get ORF Coding potential... Type: noncoding Potential: -0.999 Folding energies... MFEI: -30.016 AMFE: -0.686 GC content %: 43.770 | |
| Atauschii_AET2Gv20698800.19 AET2Gv20698800.19 |
Low | No | 2886 | OG0000052 | Get ORF Coding potential... Type: noncoding Potential: -0.976 Folding energies... MFEI: -31.327 AMFE: -0.686 GC content %: 45.634 | |
| Atauschii_AET2Gv20698800.2 AET2Gv20698800.2 |
Low | No | 1956 | OG0000052 | Get ORF Coding potential... Type: noncoding Potential: -1.125 Folding energies... MFEI: -31.150 AMFE: -0.680 GC content %: 45.808 | |
| Atauschii_AET2Gv20698800.20 AET2Gv20698800.20 |
Low | No | 2629 | OG0000052 | Get ORF Coding potential... Type: noncoding Potential: -1.055 Folding energies... MFEI: -30.730 AMFE: -0.670 GC content %: 45.835 | |
| Atauschii_AET2Gv20698800.21 AET2Gv20698800.21 |
Low | No | 3394 | OG0000052 | Get ORF Coding potential... Type: noncoding Potential: -0.990 Folding energies... MFEI: -30.866 AMFE: -0.690 GC content %: 44.726 | |
| Atauschii_AET2Gv20698800.22 AET2Gv20698800.22 |
Low | No | 2431 | OG0000052 | Get ORF Coding potential... Type: noncoding Potential: -1.048 Folding energies... MFEI: -31.802 AMFE: -0.681 GC content %: 46.689 | |
| Atauschii_AET2Gv20698800.23 AET2Gv20698800.23 |
Low | No | 2183 | OG0000052 | Get ORF Coding potential... Type: noncoding Potential: -1.037 Folding energies... MFEI: -31.796 AMFE: -0.680 GC content %: 46.725 | |
| Atauschii_AET2Gv20698800.24 AET2Gv20698800.24 |
Low | No | 2577 | OG0000052 | Get ORF Coding potential... Type: noncoding Potential: -1.151 Folding energies... MFEI: -28.619 AMFE: -0.677 GC content %: 42.297 | |
| Atauschii_AET2Gv20698800.25 AET2Gv20698800.25 |
Low | No | 3419 | OG0000052 | Get ORF Coding potential... Type: noncoding Potential: -1.134 Folding energies... MFEI: -28.347 AMFE: -0.682 GC content %: 41.591 | |
| Atauschii_AET2Gv20698800.26 AET2Gv20698800.26 |
High | No | 2331 | OG0000052 | Get ORF Coding potential... Type: noncoding Potential: -1.142 Folding energies... MFEI: -28.880 AMFE: -0.674 GC content %: 42.857 | |
| Atauschii_AET2Gv20698800.27 AET2Gv20698800.27 |
Low | No | 2517 | OG0000052 | Get ORF Coding potential... Type: noncoding Potential: -1.081 Folding energies... MFEI: -29.607 AMFE: -0.680 GC content %: 43.544 | |
| Atauschii_AET2Gv20698800.28 AET2Gv20698800.28 |
Low | No | 2226 | OG0000052 | Get ORF Coding potential... Type: noncoding Potential: -1.138 Folding energies... MFEI: -28.625 AMFE: -0.651 GC content %: 43.980 | |
| Atauschii_AET2Gv20698800.29 AET2Gv20698800.29 |
High | No | 1806 | OG0000052 | Get ORF Coding potential... Type: noncoding Potential: -1.117 Folding energies... MFEI: -29.374 AMFE: -0.675 GC content %: 43.522 | |
| Atauschii_AET2Gv20698800.3 AET2Gv20698800.3 |
Low | No | 3072 | OG0000052 | Get ORF Coding potential... Type: noncoding Potential: -0.982 Folding energies... MFEI: -31.107 AMFE: -0.690 GC content %: 45.052 | |
| Atauschii_AET2Gv20698800.30 AET2Gv20698800.30 |
High | No | 1574 | OG0000052 | Get ORF Coding potential... Type: noncoding Potential: -1.315 Folding energies... MFEI: -27.497 AMFE: -0.627 GC content %: 43.837 | |
| Atauschii_AET2Gv20698800.31 AET2Gv20698800.31 |
Low | No | 3217 | OG0000052 | Get ORF Coding potential... Type: noncoding Potential: -1.130 Folding energies... MFEI: -29.130 AMFE: -0.681 GC content %: 42.804 | |
| Atauschii_AET2Gv20698800.32 AET2Gv20698800.32 |
Low | No | 1828 | OG0000052 | Get ORF Coding potential... Type: noncoding Potential: -1.273 Folding energies... MFEI: -28.572 AMFE: -0.638 GC content %: 44.803 | |
| Atauschii_AET2Gv20698800.33 AET2Gv20698800.33 |
Low | No | 3447 | OG0000052 | Get ORF Coding potential... Type: noncoding Potential: -1.135 Folding energies... MFEI: -28.402 AMFE: -0.678 GC content %: 41.862 | |
| Atauschii_AET2Gv20698800.34 AET2Gv20698800.34 |
Low | No | 1692 | OG0000052 | Get ORF Coding potential... Type: noncoding Potential: -1.050 Folding energies... MFEI: -30.857 AMFE: -0.674 GC content %: 45.804 | |
| Atauschii_AET2Gv20698800.35 AET2Gv20698800.35 |
Low | No | 3547 | OG0000052 | Get ORF Coding potential... Type: noncoding Potential: -0.994 Folding energies... MFEI: -29.707 AMFE: -0.676 GC content %: 43.953 | |
| Atauschii_AET2Gv20698800.36 AET2Gv20698800.36 |
Low | No | 3075 | OG0000052 | Get ORF Coding potential... Type: noncoding Potential: -0.982 Folding energies... MFEI: -29.896 AMFE: -0.672 GC content %: 44.455 | |
| Atauschii_AET2Gv20698800.37 AET2Gv20698800.37 |
Low | No | 3444 | OG0000052 | Get ORF Coding potential... Type: noncoding Potential: -0.991 Folding energies... MFEI: -30.253 AMFE: -0.680 GC content %: 44.512 | |
| Atauschii_AET2Gv20698800.38 AET2Gv20698800.38 |
Low | No | 3220 | OG0000052 | Get ORF Coding potential... Type: noncoding Potential: -1.000 Folding energies... MFEI: -29.870 AMFE: -0.676 GC content %: 44.193 | |
| Atauschii_AET2Gv20698800.39 AET2Gv20698800.39 |
Low | No | 1949 | OG0000052 | Get ORF Coding potential... Type: noncoding Potential: -1.040 Folding energies... MFEI: -31.673 AMFE: -0.655 GC content %: 48.384 | |
| Atauschii_AET2Gv20698800.4 AET2Gv20698800.4 |
Low | No | 3403 | OG0000052 | Get ORF Coding potential... Type: noncoding Potential: -0.991 Folding energies... MFEI: -30.793 AMFE: -0.690 GC content %: 44.637 | |
| Atauschii_AET2Gv20698800.40 AET2Gv20698800.40 |
Low | No | 2048 | OG0000052 | Get ORF Coding potential... Type: noncoding Potential: -1.045 Folding energies... MFEI: -32.852 AMFE: -0.678 GC content %: 48.438 | |
| Atauschii_AET2Gv20698800.41 AET2Gv20698800.41 |
Low | No | 1569 | OG0000052 | Get ORF Coding potential... Type: noncoding Potential: -1.254 Folding energies... MFEI: -29.401 AMFE: -0.631 GC content %: 46.590 | |
| Atauschii_AET2Gv20698800.42 AET2Gv20698800.42 |
Low | No | 1725 | OG0000052 | Get ORF Coding potential... Type: noncoding Potential: -1.111 Folding energies... MFEI: -31.901 AMFE: -0.674 GC content %: 47.304 | |
| Atauschii_AET2Gv20698800.43 AET2Gv20698800.43 |
Low | No | 2883 | OG0000052 | Get ORF Coding potential... Type: noncoding Potential: -1.121 Folding energies... MFEI: -29.025 AMFE: -0.678 GC content %: 42.803 | |
| Atauschii_AET2Gv20698800.44 AET2Gv20698800.44 |
High | No | 887 | OG0000052 | Get ORF Coding potential... Type: noncoding Potential: -1.251 Folding energies... MFEI: -29.899 AMFE: -0.596 GC content %: 50.169 | |
| Atauschii_AET2Gv20698800.45 AET2Gv20698800.45 |
High | No | 1131 | OG0000052 | Get ORF Coding potential... Type: noncoding Potential: -1.052 Folding energies... MFEI: -26.994 AMFE: -0.635 GC content %: 42.529 | |
| Atauschii_AET2Gv20698800.5 AET2Gv20698800.5 |
Low | No | 2413 | OG0000052 | Get ORF Coding potential... Type: noncoding Potential: -1.145 Folding energies... MFEI: -29.838 AMFE: -0.685 GC content %: 43.556 | |
| Atauschii_AET2Gv20698800.6 AET2Gv20698800.6 |
Low | No | 1696 | OG0000052 | Get ORF Coding potential... Type: noncoding Potential: -1.278 Folding energies... MFEI: -32.129 AMFE: -0.693 GC content %: 46.344 | |
| Atauschii_AET2Gv20698800.8 AET2Gv20698800.8 |
Low | No | 4057 | OG0000052 | Get ORF Coding potential... Type: noncoding Potential: -1.010 Folding energies... MFEI: -29.758 AMFE: -0.688 GC content %: 43.283 | |
| Atauschii_AET2Gv20698800.9 AET2Gv20698800.9 |
Low | No | 3936 | OG0000052 | Get ORF Coding potential... Type: noncoding Potential: -1.008 Folding energies... MFEI: -29.708 AMFE: -0.686 GC content %: 43.318 |
Matches to external databases
| Transcript alias | Database | Version | Hit | Evalue |
|---|---|---|---|---|
| Atauschii_AET2Gv20698800.22 | SwissProt | 2021-04-01 | P67080 | 1.78e-7 |
| Atauschii_AET2Gv20698800.4 | SwissProt | 2021-04-01 | Q1ZXI6 | 1.09e-7 |
| Atauschii_AET2Gv20698800.18 | SwissProt | 2021-04-01 | Q1ZXI6 | 1.24e-7 |
| Atauschii_AET2Gv20698800.35 | SwissProt | 2021-04-01 | Q1ZXI6 | 1.15e-7 |
| Atauschii_AET2Gv20698800.10 | SwissProt | 2021-04-01 | P67080 | 1.68e-7 |
| Atauschii_AET2Gv20698800.3 | SwissProt | 2021-04-01 | Q1ZXI6 | 9.7e-8 |
| Atauschii_AET2Gv20698800.6 | SwissProt | 2021-04-01 | P67080 | 1.14e-7 |
| Atauschii_AET2Gv20698800.23 | SwissProt | 2021-04-01 | P67080 | 1.73e-7 |
| Atauschii_AET2Gv20698800.40 | SwissProt | 2021-04-01 | P67080 | 1.45e-7 |
| Atauschii_AET2Gv20698800.19 | SwissProt | 2021-04-01 | Q1ZXI6 | 9.0e-8 |
... further results
Gene models