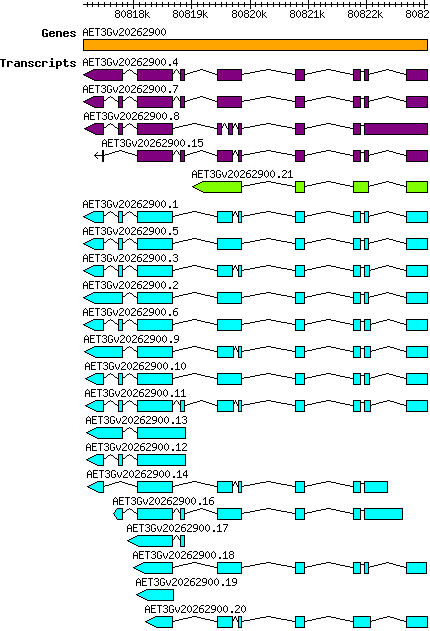
Atauschii AET3Gv20262900
From GreeNC
Gene information
| Gene alias / name | Chrom. | Start | End | Assembly | Species | Details
|
|---|---|---|---|---|---|---|
| Atauschii_AET3Gv20262900 [ AET3Gv20262900] |
3D | 80817137 | 80823045 | v4 | Aegilops tauschii (Ensembl Plants 51) | Coding? Yes |
Transcript features
This table shows how a lncRNA has been classified (high/low confidence) or whether it has been classified as pre-miRNA (go to FAQ for classification criteria). Besides, it displays the lncRNA length and sequence as well as information about the ORF, the coding potential calculated by the Coding Potential Calculator (CPC), and the folding energies calculated by RNAfold.
| Transcript alias / name | Confidence | Pre-miRNA | Length | Orthologous group | Sequence | Details
|
|---|---|---|---|---|---|---|
| Atauschii_AET3Gv20262900.1 AET3Gv20262900.1 |
Low | No | 2001 | OG0000690 | Get ORF Coding potential... Type: noncoding Potential: -0.921 Folding energies... MFEI: -34.583 AMFE: -0.671 GC content %: 51.524 | |
| Atauschii_AET3Gv20262900.10 AET3Gv20262900.10 |
Low | No | 2091 | OG0000690 | Get ORF Coding potential... Type: noncoding Potential: -1.008 Folding energies... MFEI: -33.883 AMFE: -0.670 GC content %: 50.598 | |
| Atauschii_AET3Gv20262900.11 AET3Gv20262900.11 |
Low | No | 2071 | OG0000690 | Get ORF Coding potential... Type: noncoding Potential: -0.060 Folding energies... MFEI: -34.191 AMFE: -0.674 GC content %: 50.748 | |
| Atauschii_AET3Gv20262900.12 AET3Gv20262900.12 |
Low | No | 1175 | OG0000690 | Get ORF Coding potential... Type: noncoding Potential: -1.090 Folding energies... MFEI: -33.064 AMFE: -0.703 GC content %: 47.064 | |
| Atauschii_AET3Gv20262900.13 AET3Gv20262900.13 |
Low | No | 1425 | OG0000690 | Get ORF Coding potential... Type: noncoding Potential: -1.102 Folding energies... MFEI: -31.256 AMFE: -0.707 GC content %: 44.211 | |
| Atauschii_AET3Gv20262900.14 AET3Gv20262900.14 |
Low | No | 1833 | OG0000690 | Get ORF Coding potential... Type: noncoding Potential: -1.150 Folding energies... MFEI: -32.460 AMFE: -0.690 GC content %: 47.027 | |
| Atauschii_AET3Gv20262900.16 AET3Gv20262900.16 |
Low | No | 2130 | OG0000690 | Get ORF Coding potential... Type: noncoding Potential: -0.127 Folding energies... MFEI: -31.310 AMFE: -0.682 GC content %: 45.915 | |
| Atauschii_AET3Gv20262900.17 AET3Gv20262900.17 |
Low | No | 832 | OG0000690 | Get ORF Coding potential... Type: noncoding Potential: -0.042 Folding energies... MFEI: -35.469 AMFE: -0.689 GC content %: 51.442 | |
| Atauschii_AET3Gv20262900.18 AET3Gv20262900.18 |
Low | No | 1750 | OG0000690 | Get ORF Coding potential... Type: noncoding Potential: -0.955 Folding energies... MFEI: -35.206 AMFE: -0.662 GC content %: 53.200 | |
| Atauschii_AET3Gv20262900.19 AET3Gv20262900.19 |
Low | No | 625 | OG0000690 | Get ORF Coding potential... Type: noncoding Potential: -1.033 Folding energies... MFEI: -41.408 AMFE: -0.707 GC content %: 58.560 | |
| Atauschii_AET3Gv20262900.2 AET3Gv20262900.2 |
Low | No | 2346 | OG0000690 | Get ORF Coding potential... Type: noncoding Potential: -0.950 Folding energies... MFEI: -32.664 AMFE: -0.673 GC content %: 48.508 | |
| Atauschii_AET3Gv20262900.20 AET3Gv20262900.20 |
Low | No | 1568 | OG0000690 | Get ORF Coding potential... Type: noncoding Potential: -1.020 Folding energies... MFEI: -34.566 AMFE: -0.647 GC content %: 53.444 | |
| Atauschii_AET3Gv20262900.21 AET3Gv20262900.21 |
High | No | 1602 | OG0000690 | Get ORF Coding potential... Type: noncoding Potential: -0.952 Folding energies... MFEI: -29.376 AMFE: -0.629 GC content %: 46.692 | |
| Atauschii_AET3Gv20262900.3 AET3Gv20262900.3 |
Low | No | 2027 | OG0000690 | Get ORF Coding potential... Type: noncoding Potential: -1.012 Folding energies... MFEI: -34.430 AMFE: -0.672 GC content %: 51.258 | |
| Atauschii_AET3Gv20262900.5 AET3Gv20262900.5 |
Low | No | 2099 | OG0000690 | Get ORF Coding potential... Type: noncoding Potential: -0.928 Folding energies... MFEI: -33.816 AMFE: -0.667 GC content %: 50.691 | |
| Atauschii_AET3Gv20262900.6 AET3Gv20262900.6 |
Low | No | 2140 | OG0000690 | Get ORF Coding potential... Type: noncoding Potential: -1.053 Folding energies... MFEI: -33.589 AMFE: -0.667 GC content %: 50.327 | |
| Atauschii_AET3Gv20262900.9 AET3Gv20262900.9 |
Low | No | 2277 | OG0000690 | Get ORF Coding potential... Type: noncoding Potential: -1.058 Folding energies... MFEI: -33.022 AMFE: -0.676 GC content %: 48.836 |
Matches to external databases
| Transcript alias | Database | Version | Hit | Evalue |
|---|---|---|---|---|
| Atauschii_AET3Gv20262900.12 | SwissProt | 2021-04-01 | Q8W5R1 | 1.35e-4 |
| Atauschii_AET3Gv20262900.6 | SwissProt | 2021-04-01 | P38666 | 9.34e-5 |
| Atauschii_AET3Gv20262900.19 | SwissProt | 2021-04-01 | Q8W5R1 | 5.15e-5 |
| Atauschii_AET3Gv20262900.13 | SwissProt | 2021-04-01 | Q8W5R1 | 1.87e-4 |
| Atauschii_AET3Gv20262900.9 | SwissProt | 2021-04-01 | P38666 | 2.27e-4 |
| Atauschii_AET3Gv20262900.2 | SwissProt | 2021-04-01 | P38666 | 1.04e-4 |
| Atauschii_AET3Gv20262900.14 | SwissProt | 2021-04-01 | Q8W5R1 | 4.03e-4 |
| Atauschii_AET3Gv20262900.1 | SwissProt | 2021-04-01 | Q8W5R1 | 4.55e-4 |
| Atauschii_AET3Gv20262900.20 | SwissProt | 2021-04-01 | Q8W5R1 | 3.21e-4 |
| Atauschii_AET3Gv20262900.16 | SwissProt | 2021-04-01 | Q8W5R1 | 6.68e-19 |
... further results
Gene models