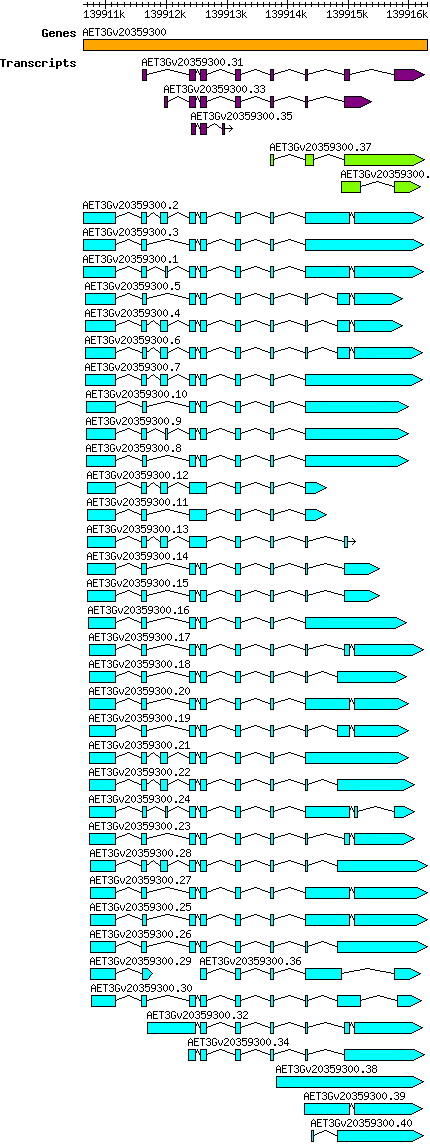
Atauschii AET3Gv20359300
From GreeNC
Gene information
| Gene alias / name | Chrom. | Start | End | Assembly | Species | Details
|
|---|---|---|---|---|---|---|
| Atauschii_AET3Gv20359300 [ AET3Gv20359300] |
3D | 139910632 | 139916304 | v4 | Aegilops tauschii (Ensembl Plants 51) | Coding? Yes |
Transcript features
This table shows how a lncRNA has been classified (high/low confidence) or whether it has been classified as pre-miRNA (go to FAQ for classification criteria). Besides, it displays the lncRNA length and sequence as well as information about the ORF, the coding potential calculated by the Coding Potential Calculator (CPC), and the folding energies calculated by RNAfold.
| Transcript alias / name | Confidence | Pre-miRNA | Length | Orthologous group | Sequence | Details
|
|---|---|---|---|---|---|---|
| Atauschii_AET3Gv20359300.1 AET3Gv20359300.1 |
Low | No | 2828 | OG0072481 | Get ORF Coding potential... Type: noncoding Potential: -0.163 Folding energies... MFEI: -28.508 AMFE: -0.669 GC content %: 42.610 | |
| Atauschii_AET3Gv20359300.10 AET3Gv20359300.10 |
Low | No | 2563 | OG0072482 | Get ORF Coding potential... Type: noncoding Potential: -0.123 Folding energies... MFEI: -27.819 AMFE: -0.663 GC content %: 41.943 | |
| Atauschii_AET3Gv20359300.11 AET3Gv20359300.11 |
Low | No | 1317 | OG0009900 | Get ORF Coding potential... Type: noncoding Potential: -0.476 Folding energies... MFEI: -30.653 AMFE: -0.619 GC content %: 49.506 | |
| Atauschii_AET3Gv20359300.12 AET3Gv20359300.12 |
Low | No | 1443 | OG0009900 | Get ORF Coding potential... Type: noncoding Potential: -1.197 Folding energies... MFEI: -30.464 AMFE: -0.629 GC content %: 48.441 | |
| Atauschii_AET3Gv20359300.13 AET3Gv20359300.13 |
Low | No | 1174 | OG0009900 | Get ORF Coding potential... Type: noncoding Potential: -1.176 Folding energies... MFEI: -33.629 AMFE: -0.653 GC content %: 51.533 | |
| Atauschii_AET3Gv20359300.14 AET3Gv20359300.14 |
Low | No | 1483 | OG0003714 | Get ORF Coding potential... Type: noncoding Potential: -0.049 Folding energies... MFEI: -29.831 AMFE: -0.623 GC content %: 47.876 | |
| Atauschii_AET3Gv20359300.15 AET3Gv20359300.15 |
Low | No | 1477 | OG0003714 | Get ORF Coding potential... Type: noncoding Potential: -0.049 Folding energies... MFEI: -29.743 AMFE: -0.622 GC content %: 47.800 | |
| Atauschii_AET3Gv20359300.16 AET3Gv20359300.16 |
Low | No | 2505 | OG0072483 | Get ORF Coding potential... Type: noncoding Potential: -0.121 Folding energies... MFEI: -27.313 AMFE: -0.655 GC content %: 41.677 | |
| Atauschii_AET3Gv20359300.17 AET3Gv20359300.17 |
Low | No | 2080 | OG0072484 | Get ORF Coding potential... Type: noncoding Potential: -0.102 Folding energies... MFEI: -28.899 AMFE: -0.659 GC content %: 43.846 | |
| Atauschii_AET3Gv20359300.18 AET3Gv20359300.18 |
Low | No | 2012 | OG0072485 | Get ORF Coding potential... Type: noncoding Potential: -0.099 Folding energies... MFEI: -28.400 AMFE: -0.655 GC content %: 43.340 | |
| Atauschii_AET3Gv20359300.19 AET3Gv20359300.19 |
Low | No | 1942 | OG0072486 | Get ORF Coding potential... Type: noncoding Potential: -0.092 Folding energies... MFEI: -28.913 AMFE: -0.664 GC content %: 43.563 | |
| Atauschii_AET3Gv20359300.2 AET3Gv20359300.2 |
Low | No | 2918 | OG0072487 | Get ORF Coding potential... Type: noncoding Potential: -0.180 Folding energies... MFEI: -28.358 AMFE: -0.671 GC content %: 42.255 | |
| Atauschii_AET3Gv20359300.20 AET3Gv20359300.20 |
Low | No | 2435 | OG0072488 | Get ORF Coding potential... Type: noncoding Potential: -0.118 Folding energies... MFEI: -27.741 AMFE: -0.666 GC content %: 41.684 | |
| Atauschii_AET3Gv20359300.21 AET3Gv20359300.21 |
Low | No | 2655 | OG0072489 | Get ORF Coding potential... Type: noncoding Potential: -0.166 Folding energies... MFEI: -27.465 AMFE: -0.665 GC content %: 41.318 | |
| Atauschii_AET3Gv20359300.22 AET3Gv20359300.22 |
Low | No | 2256 | OG0072490 | Get ORF Coding potential... Type: noncoding Potential: -0.142 Folding energies... MFEI: -28.276 AMFE: -0.657 GC content %: 43.041 | |
| Atauschii_AET3Gv20359300.23 AET3Gv20359300.23 |
Low | No | 1931 | OG0003714 | Get ORF Coding potential... Type: noncoding Potential: -0.094 Folding energies... MFEI: -29.425 AMFE: -0.658 GC content %: 44.744 | |
| Atauschii_AET3Gv20359300.24 AET3Gv20359300.24 |
Low | No | 1942 | OG0016176 | Get ORF Coding potential... Type: noncoding Potential: -0.135 Folding energies... MFEI: -29.398 AMFE: -0.656 GC content %: 44.799 | |
| Atauschii_AET3Gv20359300.25 AET3Gv20359300.25 |
Low | No | 2735 | OG0072491 | Get ORF Coding potential... Type: noncoding Potential: -0.160 Folding energies... MFEI: -27.470 AMFE: -0.670 GC content %: 40.987 | |
| Atauschii_AET3Gv20359300.26 AET3Gv20359300.26 |
Low | No | 2337 | OG0072492 | Get ORF Coding potential... Type: noncoding Potential: -0.114 Folding energies... MFEI: -28.199 AMFE: -0.668 GC content %: 42.234 | |
| Atauschii_AET3Gv20359300.27 AET3Gv20359300.27 |
Low | No | 2744 | OG0072493 | Get ORF Coding potential... Type: noncoding Potential: -0.158 Folding energies... MFEI: -27.529 AMFE: -0.671 GC content %: 40.999 | |
| Atauschii_AET3Gv20359300.28 AET3Gv20359300.28 |
Low | No | 2471 | OG0072494 | Get ORF Coding potential... Type: noncoding Potential: -0.159 Folding energies... MFEI: -28.070 AMFE: -0.668 GC content %: 42.048 | |
| Atauschii_AET3Gv20359300.29 AET3Gv20359300.29 |
Low | No | 582 | OG0016176 | Get ORF Coding potential... Type: noncoding Potential: -1.274 Folding energies... MFEI: -32.337 AMFE: -0.569 GC content %: 56.873 | |
| Atauschii_AET3Gv20359300.3 AET3Gv20359300.3 |
Low | No | 2880 | OG0072495 | Get ORF Coding potential... Type: noncoding Potential: -0.153 Folding energies... MFEI: -28.281 AMFE: -0.669 GC content %: 42.292 | |
| Atauschii_AET3Gv20359300.30 AET3Gv20359300.30 |
Low | No | 1624 | OG0004704 | Get ORF Coding potential... Type: noncoding Potential: -0.066 Folding energies... MFEI: -30.314 AMFE: -0.662 GC content %: 45.813 | |
| Atauschii_AET3Gv20359300.32 AET3Gv20359300.32 |
Low | No | 2268 | OG0072496 | Get ORF Coding potential... Type: noncoding Potential: -0.178 Folding energies... MFEI: -25.661 AMFE: -0.681 GC content %: 37.654 | |
| Atauschii_AET3Gv20359300.34 AET3Gv20359300.34 |
Low | No | 1699 | OG0003714 | Get ORF Coding potential... Type: noncoding Potential: -0.134 Folding energies... MFEI: -26.145 AMFE: -0.680 GC content %: 38.434 | |
| Atauschii_AET3Gv20359300.36 AET3Gv20359300.36 |
Low | No | 1266 | OG0004704 | Get ORF Coding potential... Type: noncoding Potential: -1.121 Folding energies... MFEI: -25.592 AMFE: -0.668 GC content %: 38.310 | |
| Atauschii_AET3Gv20359300.37 AET3Gv20359300.37 |
High | No | 1501 | OG0004704 | Get ORF Coding potential... Type: noncoding Potential: -1.147 Folding energies... MFEI: -24.437 AMFE: -0.663 GC content %: 36.842 | |
| Atauschii_AET3Gv20359300.38 AET3Gv20359300.38 |
Low | No | 2427 | OG0072497 | Get ORF Coding potential... Type: noncoding Potential: -1.202 Folding energies... MFEI: -23.988 AMFE: -0.679 GC content %: 35.311 | |
| Atauschii_AET3Gv20359300.39 AET3Gv20359300.39 |
Low | No | 1869 | OG0072498 | Get ORF Coding potential... Type: noncoding Potential: -1.175 Folding energies... MFEI: -23.884 AMFE: -0.685 GC content %: 34.885 | |
| Atauschii_AET3Gv20359300.4 AET3Gv20359300.4 |
Low | No | 2049 | OG0009900 | Get ORF Coding potential... Type: noncoding Potential: -0.132 Folding energies... MFEI: -29.527 AMFE: -0.659 GC content %: 44.802 | |
| Atauschii_AET3Gv20359300.40 AET3Gv20359300.40 |
Low | No | 1448 | OG0024986 | Get ORF Coding potential... Type: noncoding Potential: -1.142 Folding energies... MFEI: -23.978 AMFE: -0.681 GC content %: 35.221 | |
| Atauschii_AET3Gv20359300.41 AET3Gv20359300.41 |
High | No | 735 | OG0004704 | Get ORF Coding potential... Type: noncoding Potential: -1.013 Folding energies... MFEI: -25.061 AMFE: -0.685 GC content %: 36.599 | |
| Atauschii_AET3Gv20359300.5 AET3Gv20359300.5 |
Low | No | 1908 | OG0072499 | Get ORF Coding potential... Type: noncoding Potential: -0.093 Folding energies... MFEI: -29.890 AMFE: -0.661 GC content %: 45.231 | |
| Atauschii_AET3Gv20359300.6 AET3Gv20359300.6 |
Low | No | 2375 | OG0072500 | Get ORF Coding potential... Type: noncoding Potential: -0.156 Folding energies... MFEI: -28.994 AMFE: -0.663 GC content %: 43.747 | |
| Atauschii_AET3Gv20359300.7 AET3Gv20359300.7 |
Low | No | 2956 | OG0072501 | Get ORF Coding potential... Type: noncoding Potential: -0.181 Folding energies... MFEI: -27.629 AMFE: -0.661 GC content %: 41.779 | |
| Atauschii_AET3Gv20359300.8 AET3Gv20359300.8 |
Low | No | 2565 | OG0016176 | Get ORF Coding potential... Type: noncoding Potential: -0.143 Folding energies... MFEI: -27.895 AMFE: -0.664 GC content %: 42.027 | |
| Atauschii_AET3Gv20359300.9 AET3Gv20359300.9 |
Low | No | 2614 | OG0072502 | Get ORF Coding potential... Type: noncoding Potential: -0.156 Folding energies... MFEI: -27.865 AMFE: -0.661 GC content %: 42.158 |
Matches to external databases
| Transcript alias | Database | Version | Hit | Evalue |
|---|---|---|---|---|
| Atauschii_AET3Gv20359300.14 | SwissProt | 2021-04-01 | Q7F0R1 | 7.32e-14 |
| Atauschii_AET3Gv20359300.29 | SwissProt | 2021-04-01 | O80543 | 4.38e-5 |
| Atauschii_AET3Gv20359300.19 | RepBase (RepeatMasker) | 2014-01-31 | LINE/L1-Tx1 | |
| Atauschii_AET3Gv20359300.5 | RepBase (RepeatMasker) | 2014-01-31 | LINE/L1-Tx1 | |
| Atauschii_AET3Gv20359300.1 | SwissProt | 2021-04-01 | Q7F0R1 | 3.05e-13 |
| Atauschii_AET3Gv20359300.24 | SwissProt | 2021-04-01 | Q7F0R1 | 3.57e-13 |
| Atauschii_AET3Gv20359300.9 | SwissProt | 2021-04-01 | Q7F0R1 | 2.93e-13 |
| Atauschii_AET3Gv20359300.30 | RepBase (RepeatMasker) | 2014-01-31 | LINE/L1-Tx1 | |
| Atauschii_AET3Gv20359300.2 | SwissProt | 2021-04-01 | Q7F0R1 | 4.54e-13 |
| Atauschii_AET3Gv20359300.4 | SwissProt | 2021-04-01 | Q7F0R1 | 3.06e-13 |
... further results
Gene models