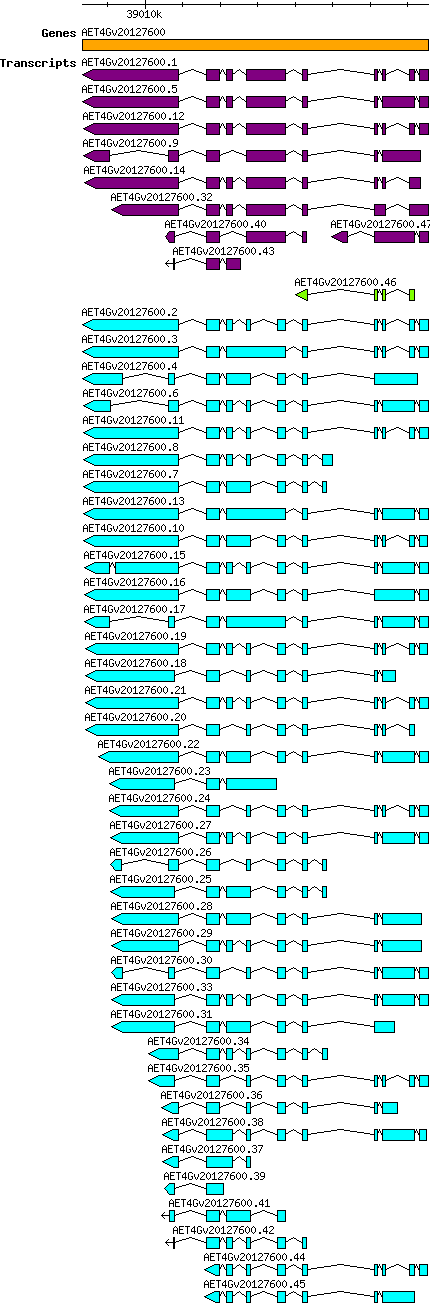
Atauschii AET4Gv20127600
From GreeNC
Gene information
| Gene alias / name | Chrom. | Start | End | Assembly | Species | Details
|
|---|---|---|---|---|---|---|
| Atauschii_AET4Gv20127600 [ AET4Gv20127600] |
4D | 39008325 | 39017560 | v4 | Aegilops tauschii (Ensembl Plants 51) | Coding? Yes |
Transcript features
This table shows how a lncRNA has been classified (high/low confidence) or whether it has been classified as pre-miRNA (go to FAQ for classification criteria). Besides, it displays the lncRNA length and sequence as well as information about the ORF, the coding potential calculated by the Coding Potential Calculator (CPC), and the folding energies calculated by RNAfold.
| Transcript alias / name | Confidence | Pre-miRNA | Length | Orthologous group | Sequence | Details
|
|---|---|---|---|---|---|---|
| Atauschii_AET4Gv20127600.10 AET4Gv20127600.10 |
Low | No | 4385 | OG0000092 | Get ORF Coding potential... Type: noncoding Potential: -1.141 Folding energies... MFEI: -29.704 AMFE: -0.686 GC content %: 43.330 | |
| Atauschii_AET4Gv20127600.11 AET4Gv20127600.11 |
Low | No | 4020 | OG0000092 | Get ORF Coding potential... Type: noncoding Potential: -1.117 Folding energies... MFEI: -30.216 AMFE: -0.682 GC content %: 44.303 | |
| Atauschii_AET4Gv20127600.13 AET4Gv20127600.13 |
Low | No | 5756 | OG0000092 | Get ORF Coding potential... Type: noncoding Potential: -1.090 Folding energies... MFEI: -28.803 AMFE: -0.689 GC content %: 41.817 | |
| Atauschii_AET4Gv20127600.15 AET4Gv20127600.15 |
Low | No | 4487 | OG0000092 | Get ORF Coding potential... Type: noncoding Potential: -1.081 Folding energies... MFEI: -29.969 AMFE: -0.687 GC content %: 43.593 | |
| Atauschii_AET4Gv20127600.16 AET4Gv20127600.16 |
Low | No | 5149 | OG0000092 | Get ORF Coding potential... Type: noncoding Potential: -1.086 Folding energies... MFEI: -29.021 AMFE: -0.686 GC content %: 42.319 | |
| Atauschii_AET4Gv20127600.17 AET4Gv20127600.17 |
Low | No | 4041 | OG0000092 | Get ORF Coding potential... Type: noncoding Potential: -1.077 Folding energies... MFEI: -30.530 AMFE: -0.689 GC content %: 44.321 | |
| Atauschii_AET4Gv20127600.18 AET4Gv20127600.18 |
Low | No | 3579 | OG0000092 | Get ORF Coding potential... Type: noncoding Potential: -1.229 Folding energies... MFEI: -29.371 AMFE: -0.673 GC content %: 43.616 | |
| Atauschii_AET4Gv20127600.19 AET4Gv20127600.19 |
Low | No | 3946 | OG0000092 | Get ORF Coding potential... Type: noncoding Potential: -1.131 Folding energies... MFEI: -30.340 AMFE: -0.684 GC content %: 44.374 | |
| Atauschii_AET4Gv20127600.2 AET4Gv20127600.2 |
Low | No | 4052 | OG0000092 | Get ORF Coding potential... Type: noncoding Potential: -1.118 Folding energies... MFEI: -30.175 AMFE: -0.683 GC content %: 44.176 | |
| Atauschii_AET4Gv20127600.20 AET4Gv20127600.20 |
Low | No | 3564 | OG0000092 | Get ORF Coding potential... Type: noncoding Potential: -1.112 Folding energies... MFEI: -29.666 AMFE: -0.681 GC content %: 43.547 | |
| Atauschii_AET4Gv20127600.21 AET4Gv20127600.21 |
Low | No | 3965 | OG0000092 | Get ORF Coding potential... Type: noncoding Potential: -1.116 Folding energies... MFEI: -30.300 AMFE: -0.680 GC content %: 44.590 | |
| Atauschii_AET4Gv20127600.22 AET4Gv20127600.22 |
Low | No | 4620 | OG0000092 | Get ORF Coding potential... Type: noncoding Potential: -1.083 Folding energies... MFEI: -28.457 AMFE: -0.683 GC content %: 41.667 | |
| Atauschii_AET4Gv20127600.23 AET4Gv20127600.23 |
Low | No | 3430 | OG0000092 | Get ORF Coding potential... Type: noncoding Potential: -1.244 Folding energies... MFEI: -24.262 AMFE: -0.653 GC content %: 37.172 | |
| Atauschii_AET4Gv20127600.24 AET4Gv20127600.24 |
Low | No | 3154 | OG0000092 | Get ORF Coding potential... Type: noncoding Potential: -1.094 Folding energies... MFEI: -29.049 AMFE: -0.683 GC content %: 42.517 | |
| Atauschii_AET4Gv20127600.25 AET4Gv20127600.25 |
Low | No | 3171 | OG0000092 | Get ORF Coding potential... Type: noncoding Potential: -1.332 Folding energies... MFEI: -27.411 AMFE: -0.663 GC content %: 41.343 | |
| Atauschii_AET4Gv20127600.26 AET4Gv20127600.26 |
Low | No | 1455 | OG0000092 | Get ORF Coding potential... Type: noncoding Potential: -1.102 Folding energies... MFEI: -33.732 AMFE: -0.680 GC content %: 49.622 | |
| Atauschii_AET4Gv20127600.27 AET4Gv20127600.27 |
Low | No | 3925 | OG0000092 | Get ORF Coding potential... Type: noncoding Potential: -1.076 Folding energies... MFEI: -28.158 AMFE: -0.680 GC content %: 41.401 | |
| Atauschii_AET4Gv20127600.28 AET4Gv20127600.28 |
Low | No | 4233 | OG0000092 | Get ORF Coding potential... Type: noncoding Potential: -1.249 Folding energies... MFEI: -26.875 AMFE: -0.676 GC content %: 39.759 | |
| Atauschii_AET4Gv20127600.29 AET4Gv20127600.29 |
Low | No | 3845 | OG0000092 | Get ORF Coding potential... Type: noncoding Potential: -1.244 Folding energies... MFEI: -27.243 AMFE: -0.677 GC content %: 40.260 | |
| Atauschii_AET4Gv20127600.3 AET4Gv20127600.3 |
Low | No | 5150 | OG0000092 | Get ORF Coding potential... Type: noncoding Potential: -1.138 Folding energies... MFEI: -28.936 AMFE: -0.682 GC content %: 42.447 | |
| Atauschii_AET4Gv20127600.30 AET4Gv20127600.30 |
Low | No | 2378 | OG0000092 | Get ORF Coding potential... Type: noncoding Potential: -1.050 Folding energies... MFEI: -31.627 AMFE: -0.693 GC content %: 45.669 | |
| Atauschii_AET4Gv20127600.31 AET4Gv20127600.31 |
Low | No | 3564 | OG0000092 | Get ORF Coding potential... Type: noncoding Potential: -1.343 Folding energies... MFEI: -26.667 AMFE: -0.666 GC content %: 40.011 | |
| Atauschii_AET4Gv20127600.33 AET4Gv20127600.33 |
Low | No | 3796 | OG0000092 | Get ORF Coding potential... Type: noncoding Potential: -1.075 Folding energies... MFEI: -28.322 AMFE: -0.682 GC content %: 41.544 | |
| Atauschii_AET4Gv20127600.34 AET4Gv20127600.34 |
Low | No | 1884 | OG0000092 | Get ORF Coding potential... Type: noncoding Potential: -1.099 Folding energies... MFEI: -30.711 AMFE: -0.685 GC content %: 44.851 | |
| Atauschii_AET4Gv20127600.35 AET4Gv20127600.35 |
Low | No | 2009 | OG0000092 | Get ORF Coding potential... Type: noncoding Potential: -1.059 Folding energies... MFEI: -31.518 AMFE: -0.700 GC content %: 45.047 | |
| Atauschii_AET4Gv20127600.36 AET4Gv20127600.36 |
Low | No | 1696 | OG0000092 | Get ORF Coding potential... Type: noncoding Potential: -1.130 Folding energies... MFEI: -30.554 AMFE: -0.684 GC content %: 44.693 | |
| Atauschii_AET4Gv20127600.37 AET4Gv20127600.37 |
Low | No | 1214 | OG0000092 | Get ORF Coding potential... Type: noncoding Potential: -1.389 Folding energies... MFEI: -24.992 AMFE: -0.635 GC content %: 39.374 | |
| Atauschii_AET4Gv20127600.38 AET4Gv20127600.38 |
Low | No | 2661 | OG0000092 | Get ORF Coding potential... Type: noncoding Potential: -1.205 Folding energies... MFEI: -29.342 AMFE: -0.683 GC content %: 42.954 | |
| Atauschii_AET4Gv20127600.39 AET4Gv20127600.39 |
Low | No | 727 | OG0000092 | Get ORF Coding potential... Type: noncoding Potential: -1.182 Folding energies... MFEI: -25.447 AMFE: -0.658 GC content %: 38.652 | |
| Atauschii_AET4Gv20127600.4 AET4Gv20127600.4 |
Low | No | 3693 | OG0000092 | Get ORF Coding potential... Type: noncoding Potential: -1.242 Folding energies... MFEI: -29.892 AMFE: -0.686 GC content %: 43.596 | |
| Atauschii_AET4Gv20127600.41 AET4Gv20127600.41 |
Low | No | 1371 | OG0000092 | Get ORF Coding potential... Type: noncoding Potential: -1.211 Folding energies... MFEI: -27.892 AMFE: -0.644 GC content %: 43.326 | |
| Atauschii_AET4Gv20127600.42 AET4Gv20127600.42 |
Low | No | 984 | OG0000092 | Get ORF Coding potential... Type: noncoding Potential: -1.137 Folding energies... MFEI: -32.632 AMFE: -0.650 GC content %: 50.203 | |
| Atauschii_AET4Gv20127600.44 AET4Gv20127600.44 |
Low | No | 1512 | OG0000092 | Get ORF Coding potential... Type: noncoding Potential: -1.022 Folding energies... MFEI: -33.671 AMFE: -0.688 GC content %: 48.942 | |
| Atauschii_AET4Gv20127600.45 AET4Gv20127600.45 |
Low | No | 1900 | OG0000092 | Get ORF Coding potential... Type: noncoding Potential: -1.160 Folding energies... MFEI: -29.147 AMFE: -0.674 GC content %: 43.263 | |
| Atauschii_AET4Gv20127600.46 AET4Gv20127600.46 |
High | No | 600 | OG0000092 | Get ORF Coding potential... Type: noncoding Potential: -0.752 Folding energies... MFEI: -26.850 AMFE: -0.590 GC content %: 45.500 | |
| Atauschii_AET4Gv20127600.6 AET4Gv20127600.6 |
Low | No | 3081 | OG0000092 | Get ORF Coding potential... Type: noncoding Potential: -1.065 Folding energies... MFEI: -32.438 AMFE: -0.692 GC content %: 46.900 | |
| Atauschii_AET4Gv20127600.7 AET4Gv20127600.7 |
Low | No | 3985 | OG0000092 | Get ORF Coding potential... Type: noncoding Potential: -1.188 Folding energies... MFEI: -29.345 AMFE: -0.677 GC content %: 43.363 | |
| Atauschii_AET4Gv20127600.8 AET4Gv20127600.8 |
Low | No | 3775 | OG0000092 | Get ORF Coding potential... Type: noncoding Potential: -1.289 Folding energies... MFEI: -31.128 AMFE: -0.682 GC content %: 45.616 |
Matches to external databases
| Transcript alias | Database | Version | Hit | Evalue |
|---|---|---|---|---|
| Atauschii_AET4Gv20127600.39 | SwissProt | 2021-04-01 | Q940Y1 | 1.21e-8 |
| Atauschii_AET4Gv20127600.23 | SwissProt | 2021-04-01 | Q940Y1 | 4.85e-7 |
| Atauschii_AET4Gv20127600.34 | SwissProt | 2021-04-01 | Q940Y1 | 2.12e-7 |
| Atauschii_AET4Gv20127600.19 | SwissProt | 2021-04-01 | Q940Y1 | 5.76e-7 |
| Atauschii_AET4Gv20127600.29 | SwissProt | 2021-04-01 | Q940Y1 | 5.58e-7 |
| Atauschii_AET4Gv20127600.6 | SwissProt | 2021-04-01 | Q940Y1 | 4.23e-7 |
| Atauschii_AET4Gv20127600.13 | SwissProt | 2021-04-01 | Q940Y1 | 8.95e-7 |
| Atauschii_AET4Gv20127600.4 | SwissProt | 2021-04-01 | Q940Y1 | 5.32e-7 |
| Atauschii_AET4Gv20127600.24 | SwissProt | 2021-04-01 | Q940Y1 | 1.94e-7 |
| Atauschii_AET4Gv20127600.35 | SwissProt | 2021-04-01 | Q940Y1 | 1.04e-7 |
... further results
Gene models