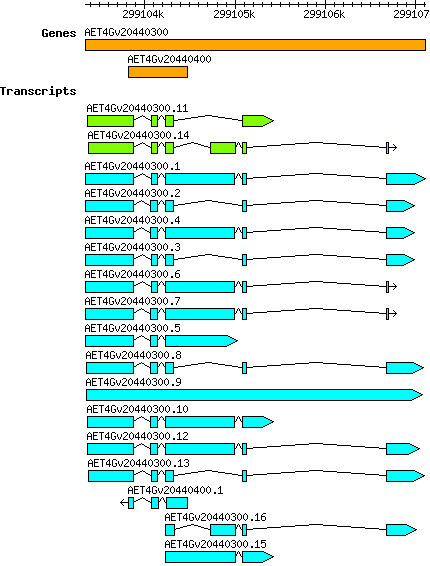
Atauschii AET4Gv20440300
From GreeNC
Gene information
| Gene alias / name | Chrom. | Start | End | Assembly | Species | Details
|
|---|---|---|---|---|---|---|
| Atauschii_AET4Gv20440300 [ AET4Gv20440300] |
4D | 299103340 | 299107110 | v4 | Aegilops tauschii (Ensembl Plants 51) | Coding? No |
Transcript features
This table shows how a lncRNA has been classified (high/low confidence) or whether it has been classified as pre-miRNA (go to FAQ for classification criteria). Besides, it displays the lncRNA length and sequence as well as information about the ORF, the coding potential calculated by the Coding Potential Calculator (CPC), and the folding energies calculated by RNAfold.
| Transcript alias / name | Confidence | Pre-miRNA | Length | Orthologous group | Sequence | Details
|
|---|---|---|---|---|---|---|
| Atauschii_AET4Gv20440300.1 AET4Gv20440300.1 |
Low | No | 1843 | OG0000024 | Get ORF Coding potential... Type: noncoding Potential: -0.978 Folding energies... MFEI: -28.171 AMFE: -0.579 GC content %: 48.671 | |
| Atauschii_AET4Gv20440300.10 AET4Gv20440300.10 |
Low | No | 1692 | OG0000024 | Get ORF Coding potential... Type: noncoding Potential: -0.965 Folding energies... MFEI: -27.299 AMFE: -0.553 GC content %: 49.350 | |
| Atauschii_AET4Gv20440300.11 AET4Gv20440300.11 |
High | No | 1007 | OG0000024 | Get ORF Coding potential... Type: noncoding Potential: -1.085 Folding energies... MFEI: -27.388 AMFE: -0.501 GC content %: 54.717 | |
| Atauschii_AET4Gv20440300.12 AET4Gv20440300.12 |
Low | No | 1760 | OG0000024 | Get ORF Coding potential... Type: noncoding Potential: -0.971 Folding energies... MFEI: -28.699 AMFE: -0.580 GC content %: 49.489 | |
| Atauschii_AET4Gv20440300.13 AET4Gv20440300.13 |
Low | No | 1123 | OG0000024 | Get ORF Coding potential... Type: noncoding Potential: -1.137 Folding energies... MFEI: -29.920 AMFE: -0.562 GC content %: 53.250 | |
| Atauschii_AET4Gv20440300.14 AET4Gv20440300.14 |
High | No | 989 | OG0000024 | Get ORF Coding potential... Type: noncoding Potential: -0.802 Folding energies... MFEI: -32.285 AMFE: -0.545 GC content %: 59.252 | |
| Atauschii_AET4Gv20440300.15 AET4Gv20440300.15 |
Low | No | 1106 | OG0000024 | Get ORF Coding potential... Type: noncoding Potential: -0.884 Folding energies... MFEI: -24.991 AMFE: -0.607 GC content %: 41.139 | |
| Atauschii_AET4Gv20440300.16 AET4Gv20440300.16 |
Low | No | 732 | OG0000024 | Get ORF Coding potential... Type: noncoding Potential: -0.734 Folding energies... MFEI: -29.577 AMFE: -0.650 GC content %: 45.492 | |
| Atauschii_AET4Gv20440300.2 AET4Gv20440300.2 |
Low | No | 1042 | OG0000024 | Get ORF Coding potential... Type: noncoding Potential: -1.083 Folding energies... MFEI: -30.749 AMFE: -0.555 GC content %: 55.374 | |
| Atauschii_AET4Gv20440300.3 AET4Gv20440300.3 |
Low | No | 1032 | OG0000024 | Get ORF Coding potential... Type: noncoding Potential: -1.131 Folding energies... MFEI: -30.659 AMFE: -0.552 GC content %: 55.523 | |
| Atauschii_AET4Gv20440300.4 AET4Gv20440300.4 |
Low | No | 1720 | OG0000024 | Get ORF Coding potential... Type: noncoding Potential: -0.967 Folding energies... MFEI: -28.977 AMFE: -0.581 GC content %: 49.884 | |
| Atauschii_AET4Gv20440300.5 AET4Gv20440300.5 |
Low | No | 1409 | OG0000024 | Get ORF Coding potential... Type: noncoding Potential: -0.893 Folding energies... MFEI: -28.985 AMFE: -0.557 GC content %: 52.023 | |
| Atauschii_AET4Gv20440300.6 AET4Gv20440300.6 |
Low | No | 1424 | OG0000024 | Get ORF Coding potential... Type: noncoding Potential: -0.935 Folding energies... MFEI: -28.820 AMFE: -0.550 GC content %: 52.388 | |
| Atauschii_AET4Gv20440300.7 AET4Gv20440300.7 |
Low | No | 1441 | OG0000024 | Get ORF Coding potential... Type: noncoding Potential: -0.937 Folding energies... MFEI: -29.008 AMFE: -0.555 GC content %: 52.255 | |
| Atauschii_AET4Gv20440300.8 AET4Gv20440300.8 |
Low | No | 1138 | OG0000024 | Get ORF Coding potential... Type: noncoding Potential: -1.139 Folding energies... MFEI: -29.886 AMFE: -0.560 GC content %: 53.339 | |
| Atauschii_AET4Gv20440300.9 AET4Gv20440300.9 |
Low | No | 3713 | OG0000024 | Get ORF Coding potential... Type: noncoding Potential: -1.096 Folding energies... MFEI: -24.899 AMFE: -0.589 GC content %: 42.284 |
Matches to external databases
| Transcript alias | Database | Version | Hit | Evalue |
|---|---|---|---|---|
| Atauschii_AET4Gv20440300.12 | RepBase (RepeatMasker) | 2014-01-31 | DNA/CMC-EnSpm; LTR/Gypsy | |
| Atauschii_AET4Gv20440300.9 | RepBase (RepeatMasker) | 2014-01-31 | DNA/CMC-EnSpm; LTR/Gypsy | |
| Atauschii_AET4Gv20440300.4 | RepBase (RepeatMasker) | 2014-01-31 | DNA/CMC-EnSpm; LTR/Gypsy | |
| Atauschii_AET4Gv20440300.13 | RepBase (RepeatMasker) | 2014-01-31 | DNA/CMC-EnSpm | |
| Atauschii_AET4Gv20440300.5 | RepBase (RepeatMasker) | 2014-01-31 | LTR/Gypsy | |
| Atauschii_AET4Gv20440300.15 | RepBase (RepeatMasker) | 2014-01-31 | LTR/Gypsy | |
| Atauschii_AET4Gv20440300.6 | RepBase (RepeatMasker) | 2014-01-31 | LTR/Gypsy | |
| Atauschii_AET4Gv20440300.16 | RepBase (RepeatMasker) | 2014-01-31 | DNA/CMC-EnSpm | |
| Atauschii_AET4Gv20440300.1 | RepBase (RepeatMasker) | 2014-01-31 | DNA/CMC-EnSpm; LTR/Gypsy | |
| Atauschii_AET4Gv20440300.7 | RepBase (RepeatMasker) | 2014-01-31 | LTR/Gypsy |
... further results
Gene models