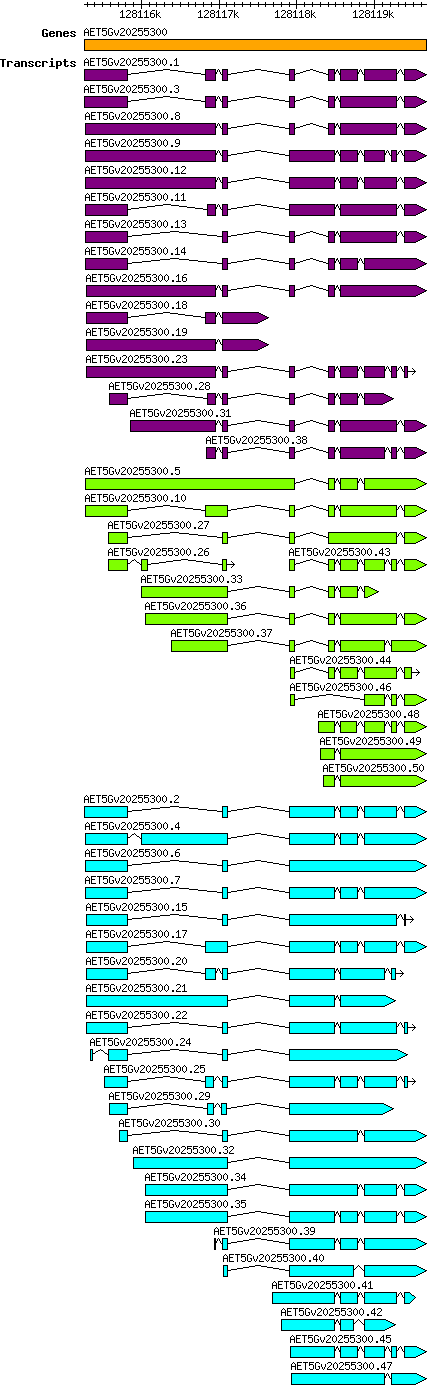
Atauschii AET5Gv20255300
From GreeNC
Gene information
| Gene alias / name | Chrom. | Start | End | Assembly | Species | Details
|
|---|---|---|---|---|---|---|
| Atauschii_AET5Gv20255300 [ AET5Gv20255300] |
5D | 128115266 | 128119667 | v4 | Aegilops tauschii (Ensembl Plants 51) | Coding? Yes |
Transcript features
This table shows how a lncRNA has been classified (high/low confidence) or whether it has been classified as pre-miRNA (go to FAQ for classification criteria). Besides, it displays the lncRNA length and sequence as well as information about the ORF, the coding potential calculated by the Coding Potential Calculator (CPC), and the folding energies calculated by RNAfold.
| Transcript alias / name | Confidence | Pre-miRNA | Length | Orthologous group | Sequence | Details
|
|---|---|---|---|---|---|---|
| Atauschii_AET5Gv20255300.10 AET5Gv20255300.10 |
High | No | 1960 | OG0000145 | Get ORF Coding potential... Type: noncoding Potential: -1.062 Folding energies... MFEI: -30.357 AMFE: -0.575 GC content %: 52.755 | |
| Atauschii_AET5Gv20255300.15 AET5Gv20255300.15 |
Low | Yes | 1979 | OG0000145 | Get ORF Coding potential... Type: noncoding Potential: -1.063 Folding energies... MFEI: -29.909 AMFE: -0.619 GC content %: 48.307 | |
| Atauschii_AET5Gv20255300.17 AET5Gv20255300.17 |
Low | Yes | 2286 | OG0000145 | Get ORF Coding potential... Type: noncoding Potential: -1.081 Folding energies... MFEI: -31.378 AMFE: -0.626 GC content %: 50.087 | |
| Atauschii_AET5Gv20255300.2 AET5Gv20255300.2 |
Low | Yes | 2099 | OG0000145 | Get ORF Coding potential... Type: noncoding Potential: -1.071 Folding energies... MFEI: -31.363 AMFE: -0.623 GC content %: 50.357 | |
| Atauschii_AET5Gv20255300.20 AET5Gv20255300.20 |
Low | Yes | 1903 | OG0000145 | Get ORF Coding potential... Type: noncoding Potential: -1.010 Folding energies... MFEI: -31.088 AMFE: -0.624 GC content %: 49.816 | |
| Atauschii_AET5Gv20255300.21 AET5Gv20255300.21 |
Low | Yes | 3096 | OG0000145 | Get ORF Coding potential... Type: noncoding Potential: -1.111 Folding energies... MFEI: -28.085 AMFE: -0.634 GC content %: 44.315 | |
| Atauschii_AET5Gv20255300.22 AET5Gv20255300.22 |
Low | Yes | 1922 | OG0000145 | Get ORF Coding potential... Type: noncoding Potential: -1.059 Folding energies... MFEI: -30.598 AMFE: -0.623 GC content %: 49.116 | |
| Atauschii_AET5Gv20255300.24 AET5Gv20255300.24 |
Low | Yes | 1845 | OG0000145 | Get ORF Coding potential... Type: noncoding Potential: -1.089 Folding energies... MFEI: -29.106 AMFE: -0.675 GC content %: 43.089 | |
| Atauschii_AET5Gv20255300.25 AET5Gv20255300.25 |
Low | Yes | 1690 | OG0000145 | Get ORF Coding potential... Type: noncoding Potential: -1.024 Folding energies... MFEI: -30.544 AMFE: -0.658 GC content %: 46.450 | |
| Atauschii_AET5Gv20255300.26 AET5Gv20255300.26 |
High | No | 376 | OG0076194 | Get ORF Coding potential... Type: noncoding Potential: -1.061 Folding energies... MFEI: -25.612 AMFE: -0.463 GC content %: 55.319 | |
| Atauschii_AET5Gv20255300.27 AET5Gv20255300.27 |
High | No | 1530 | OG0000145 | Get ORF Coding potential... Type: noncoding Potential: -1.263 Folding energies... MFEI: -28.765 AMFE: -0.610 GC content %: 47.124 | |
| Atauschii_AET5Gv20255300.29 AET5Gv20255300.29 |
Low | Yes | 1698 | OG0000145 | Get ORF Coding potential... Type: noncoding Potential: -1.241 Folding energies... MFEI: -28.975 AMFE: -0.676 GC content %: 42.874 | |
| Atauschii_AET5Gv20255300.30 AET5Gv20255300.30 |
Low | Yes | 1833 | OG0000145 | Get ORF Coding potential... Type: noncoding Potential: -1.089 Folding energies... MFEI: -29.165 AMFE: -0.690 GC content %: 42.280 | |
| Atauschii_AET5Gv20255300.32 AET5Gv20255300.32 |
Low | Yes | 2965 | OG0000145 | Get ORF Coding potential... Type: noncoding Potential: -1.137 Folding energies... MFEI: -27.194 AMFE: -0.690 GC content %: 39.427 | |
| Atauschii_AET5Gv20255300.33 AET5Gv20255300.33 |
High | No | 1650 | OG0000145 | Get ORF Coding potential... Type: noncoding Potential: -1.269 Folding energies... MFEI: -25.345 AMFE: -0.637 GC content %: 39.818 | |
| Atauschii_AET5Gv20255300.34 AET5Gv20255300.34 |
Low | Yes | 2616 | OG0000145 | Get ORF Coding potential... Type: noncoding Potential: -1.265 Folding energies... MFEI: -27.726 AMFE: -0.691 GC content %: 40.099 | |
| Atauschii_AET5Gv20255300.35 AET5Gv20255300.35 |
Low | Yes | 2541 | OG0000145 | Get ORF Coding potential... Type: noncoding Potential: -1.264 Folding energies... MFEI: -27.946 AMFE: -0.691 GC content %: 40.417 | |
| Atauschii_AET5Gv20255300.36 AET5Gv20255300.36 |
High | No | 2211 | OG0000145 | Get ORF Coding potential... Type: noncoding Potential: -1.276 Folding energies... MFEI: -26.495 AMFE: -0.642 GC content %: 41.294 | |
| Atauschii_AET5Gv20255300.37 AET5Gv20255300.37 |
High | No | 1886 | OG0000145 | Get ORF Coding potential... Type: noncoding Potential: -1.275 Folding energies... MFEI: -26.972 AMFE: -0.643 GC content %: 41.941 | |
| Atauschii_AET5Gv20255300.39 AET5Gv20255300.39 |
Low | Yes | 1654 | OG0000145 | Get ORF Coding potential... Type: noncoding Potential: -1.236 Folding energies... MFEI: -30.073 AMFE: -0.717 GC content %: 41.959 | |
| Atauschii_AET5Gv20255300.4 AET5Gv20255300.4 |
Low | Yes | 3233 | OG0000145 | Get ORF Coding potential... Type: noncoding Potential: -1.114 Folding energies... MFEI: -28.704 AMFE: -0.631 GC content %: 45.469 | |
| Atauschii_AET5Gv20255300.40 AET5Gv20255300.40 |
Low | Yes | 1662 | OG0000145 | Get ORF Coding potential... Type: noncoding Potential: -1.237 Folding energies... MFEI: -29.555 AMFE: -0.709 GC content %: 41.697 | |
| Atauschii_AET5Gv20255300.41 AET5Gv20255300.41 |
Low | Yes | 1573 | OG0000145 | Get ORF Coding potential... Type: noncoding Potential: -1.007 Folding energies... MFEI: -31.132 AMFE: -0.722 GC content %: 43.102 | |
| Atauschii_AET5Gv20255300.42 AET5Gv20255300.42 |
Low | Yes | 1254 | OG0000145 | Get ORF Coding potential... Type: noncoding Potential: -0.990 Folding energies... MFEI: -30.311 AMFE: -0.754 GC content %: 40.191 | |
| Atauschii_AET5Gv20255300.43 AET5Gv20255300.43 |
High | No | 954 | OG0000145 | Get ORF Coding potential... Type: noncoding Potential: -1.238 Folding energies... MFEI: -29.853 AMFE: -0.627 GC content %: 47.589 | |
| Atauschii_AET5Gv20255300.44 AET5Gv20255300.44 |
High | No | 858 | OG0000145 | Get ORF Coding potential... Type: noncoding Potential: -1.281 Folding energies... MFEI: -28.776 AMFE: -0.623 GC content %: 46.154 | |
| Atauschii_AET5Gv20255300.45 AET5Gv20255300.45 |
Low | Yes | 1378 | OG0000145 | Get ORF Coding potential... Type: noncoding Potential: -1.258 Folding energies... MFEI: -31.328 AMFE: -0.724 GC content %: 43.251 | |
| Atauschii_AET5Gv20255300.46 AET5Gv20255300.46 |
High | No | 653 | OG0000145 | Get ORF Coding potential... Type: noncoding Potential: -1.207 Folding energies... MFEI: -27.213 AMFE: -0.598 GC content %: 45.482 | |
| Atauschii_AET5Gv20255300.47 AET5Gv20255300.47 |
Low | Yes | 1643 | OG0000145 | Get ORF Coding potential... Type: noncoding Potential: -1.341 Folding energies... MFEI: -29.203 AMFE: -0.712 GC content %: 41.023 | |
| Atauschii_AET5Gv20255300.48 AET5Gv20255300.48 |
High | No | 999 | OG0000145 | Get ORF Coding potential... Type: noncoding Potential: -1.240 Folding energies... MFEI: -28.529 AMFE: -0.617 GC content %: 46.246 | |
| Atauschii_AET5Gv20255300.49 AET5Gv20255300.49 |
High | No | 1282 | OG0000145 | Get ORF Coding potential... Type: noncoding Potential: -1.322 Folding energies... MFEI: -27.402 AMFE: -0.632 GC content %: 43.370 | |
| Atauschii_AET5Gv20255300.5 AET5Gv20255300.5 |
High | No | 3789 | OG0000145 | Get ORF Coding potential... Type: noncoding Potential: -1.126 Folding energies... MFEI: -28.334 AMFE: -0.608 GC content %: 46.609 | |
| Atauschii_AET5Gv20255300.50 AET5Gv20255300.50 |
High | No | 1251 | OG0000145 | Get ORF Coding potential... Type: noncoding Potential: -1.320 Folding energies... MFEI: -27.426 AMFE: -0.628 GC content %: 43.645 | |
| Atauschii_AET5Gv20255300.6 AET5Gv20255300.6 |
Low | Yes | 2366 | OG0000145 | Get ORF Coding potential... Type: noncoding Potential: -1.084 Folding energies... MFEI: -30.262 AMFE: -0.630 GC content %: 48.056 | |
| Atauschii_AET5Gv20255300.7 AET5Gv20255300.7 |
Low | Yes | 2188 | OG0000145 | Get ORF Coding potential... Type: noncoding Potential: -1.076 Folding energies... MFEI: -30.850 AMFE: -0.623 GC content %: 49.543 |
Matches to external databases
| Transcript alias | Database | Version | Hit | Evalue |
|---|---|---|---|---|
| Atauschii_AET5Gv20255300.15 | RepBase (RepeatMasker) | 2014-01-31 | DNA/CMC-EnSpm; DNA/TcMar-Stowaway | |
| Atauschii_AET5Gv20255300.41 | RepBase (RepeatMasker) | 2014-01-31 | DNA/CMC-EnSpm; DNA/TcMar-Stowaway | |
| Atauschii_AET5Gv20255300.24 | Rfam | 14.6 | MIR1122 | 3.2e-13 |
| Atauschii_AET5Gv20255300.42 | Rfam | 14.6 | MIR1122 | 2.2e-13 |
| Atauschii_AET5Gv20255300.34 | RepBase (RepeatMasker) | 2014-01-31 | DNA/CMC-EnSpm; DNA/TcMar-Stowaway | |
| Atauschii_AET5Gv20255300.17 | Rfam | 14.6 | MIR1122 | 3.9e-13 |
| Atauschii_AET5Gv20255300.24 | RepBase (RepeatMasker) | 2014-01-31 | DNA/CMC-EnSpm; DNA/TcMar-Stowaway | |
| Atauschii_AET5Gv20255300.35 | Rfam | 14.6 | MIR1122 | 4.4e-13 |
| Atauschii_AET5Gv20255300.17 | RepBase (RepeatMasker) | 2014-01-31 | DNA/CMC-EnSpm; DNA/TcMar-Stowaway | |
| Atauschii_AET5Gv20255300.42 | RepBase (RepeatMasker) | 2014-01-31 | DNA/CMC-EnSpm; DNA/TcMar-Stowaway |
... further results
Gene models