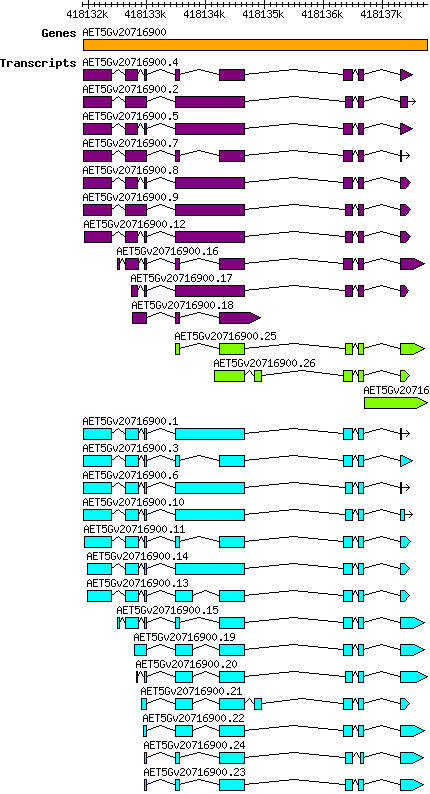
Atauschii AET5Gv20716900
From GreeNC
Gene information
| Gene alias / name | Chrom. | Start | End | Assembly | Species | Details
|
|---|---|---|---|---|---|---|
| Atauschii_AET5Gv20716900 [ AET5Gv20716900] |
5D | 418131920 | 418137769 | v4 | Aegilops tauschii (Ensembl Plants 51) | Coding? Yes |
Transcript features
This table shows how a lncRNA has been classified (high/low confidence) or whether it has been classified as pre-miRNA (go to FAQ for classification criteria). Besides, it displays the lncRNA length and sequence as well as information about the ORF, the coding potential calculated by the Coding Potential Calculator (CPC), and the folding energies calculated by RNAfold.
| Transcript alias / name | Confidence | Pre-miRNA | Length | Orthologous group | Sequence | Details
|
|---|---|---|---|---|---|---|
| Atauschii_AET5Gv20716900.1 AET5Gv20716900.1 |
Low | No | 2196 | OG0000066 | Get ORF Coding potential... Type: noncoding Potential: -0.427 Folding energies... MFEI: -31.730 AMFE: -0.659 GC content %: 48.133 | |
| Atauschii_AET5Gv20716900.10 AET5Gv20716900.10 |
Low | No | 2199 | OG0000066 | Get ORF Coding potential... Type: noncoding Potential: -0.427 Folding energies... MFEI: -31.810 AMFE: -0.659 GC content %: 48.249 | |
| Atauschii_AET5Gv20716900.11 AET5Gv20716900.11 |
Low | No | 1656 | OG0000066 | Get ORF Coding potential... Type: noncoding Potential: -0.342 Folding energies... MFEI: -35.236 AMFE: -0.651 GC content %: 54.167 | |
| Atauschii_AET5Gv20716900.13 AET5Gv20716900.13 |
Low | No | 1811 | OG0000066 | Get ORF Coding potential... Type: noncoding Potential: -0.396 Folding energies... MFEI: -33.363 AMFE: -0.652 GC content %: 51.187 | |
| Atauschii_AET5Gv20716900.14 AET5Gv20716900.14 |
Low | No | 2268 | OG0000066 | Get ORF Coding potential... Type: noncoding Potential: -0.431 Folding energies... MFEI: -31.314 AMFE: -0.661 GC content %: 47.354 | |
| Atauschii_AET5Gv20716900.15 AET5Gv20716900.15 |
Low | No | 1453 | OG0000066 | Get ORF Coding potential... Type: noncoding Potential: -0.318 Folding energies... MFEI: -29.780 AMFE: -0.632 GC content %: 47.144 | |
| Atauschii_AET5Gv20716900.19 AET5Gv20716900.19 |
Low | No | 1588 | OG0000066 | Get ORF Coding potential... Type: noncoding Potential: -0.276 Folding energies... MFEI: -26.751 AMFE: -0.613 GC content %: 43.640 | |
| Atauschii_AET5Gv20716900.20 AET5Gv20716900.20 |
Low | No | 1443 | OG0000066 | Get ORF Coding potential... Type: noncoding Potential: -0.298 Folding energies... MFEI: -26.459 AMFE: -0.612 GC content %: 43.243 | |
| Atauschii_AET5Gv20716900.21 AET5Gv20716900.21 |
Low | No | 1341 | OG0000066 | Get ORF Coding potential... Type: noncoding Potential: -0.397 Folding energies... MFEI: -27.069 AMFE: -0.623 GC content %: 43.475 | |
| Atauschii_AET5Gv20716900.22 AET5Gv20716900.22 |
Low | No | 1450 | OG0000066 | Get ORF Coding potential... Type: noncoding Potential: -0.348 Folding energies... MFEI: -26.517 AMFE: -0.610 GC content %: 43.448 | |
| Atauschii_AET5Gv20716900.23 AET5Gv20716900.23 |
Low | No | 1130 | OG0000066 | Get ORF Coding potential... Type: noncoding Potential: -1.090 Folding energies... MFEI: -27.053 AMFE: -0.598 GC content %: 45.221 | |
| Atauschii_AET5Gv20716900.24 AET5Gv20716900.24 |
Low | No | 1082 | OG0000066 | Get ORF Coding potential... Type: noncoding Potential: -1.084 Folding energies... MFEI: -26.229 AMFE: -0.586 GC content %: 44.732 | |
| Atauschii_AET5Gv20716900.25 AET5Gv20716900.25 |
High | No | 1106 | OG0000066 | Get ORF Coding potential... Type: noncoding Potential: -1.087 Folding energies... MFEI: -26.700 AMFE: -0.594 GC content %: 44.937 | |
| Atauschii_AET5Gv20716900.26 AET5Gv20716900.26 |
High | No | 1044 | OG0000066 | Get ORF Coding potential... Type: noncoding Potential: -1.129 Folding energies... MFEI: -27.337 AMFE: -0.623 GC content %: 43.870 | |
| Atauschii_AET5Gv20716900.27 AET5Gv20716900.27 |
High | No | 1080 | OG0000066 | Get ORF Coding potential... Type: noncoding Potential: -1.039 Folding energies... MFEI: -24.148 AMFE: -0.609 GC content %: 39.630 | |
| Atauschii_AET5Gv20716900.3 AET5Gv20716900.3 |
Low | No | 1696 | OG0000066 | Get ORF Coding potential... Type: noncoding Potential: -0.345 Folding energies... MFEI: -34.941 AMFE: -0.645 GC content %: 54.186 | |
| Atauschii_AET5Gv20716900.6 AET5Gv20716900.6 |
Low | No | 2151 | OG0000066 | Get ORF Coding potential... Type: noncoding Potential: -0.425 Folding energies... MFEI: -31.683 AMFE: -0.659 GC content %: 48.071 |
Matches to external databases
| Transcript alias | Database | Version | Hit | Evalue |
|---|---|---|---|---|
| Atauschii_AET5Gv20716900.10 | SwissProt | 2021-04-01 | Q9ZT42 | 4.18e-4 |
| Atauschii_AET5Gv20716900.3 | SwissProt | 2021-04-01 | Q9ZT42 | 7.81e-4 |
| Atauschii_AET5Gv20716900.20 | SwissProt | 2021-04-01 | Q9ZT42 | 1.48e-12 |
| Atauschii_AET5Gv20716900.11 | SwissProt | 2021-04-01 | Q9ZT42 | 7.56e-4 |
| Atauschii_AET5Gv20716900.6 | SwissProt | 2021-04-01 | Q9ZT42 | 4.06e-4 |
| Atauschii_AET5Gv20716900.21 | SwissProt | 2021-04-01 | Q9ZT42 | 7.94e-12 |
| Atauschii_AET5Gv20716900.13 | SwissProt | 2021-04-01 | Q9ZT42 | 3.23e-4 |
| Atauschii_AET5Gv20716900.22 | SwissProt | 2021-04-01 | Q9ZT42 | 9.35e-12 |
| Atauschii_AET5Gv20716900.14 | SwissProt | 2021-04-01 | Q9ZT42 | 4.35e-4 |
| Atauschii_AET5Gv20716900.23 | SwissProt | 2021-04-01 | Q9ZT42 | 6.41e-10 |
... further results
Gene models