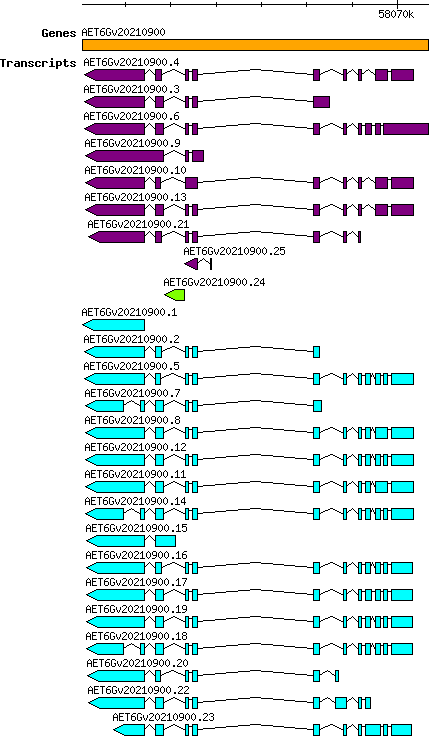
Atauschii AET6Gv20210900
From GreeNC
Gene information
| Gene alias / name | Chrom. | Start | End | Assembly | Species | Details
|
|---|---|---|---|---|---|---|
| Atauschii_AET6Gv20210900 [ AET6Gv20210900] |
6D | 58063063 | 58070669 | v4 | Aegilops tauschii (Ensembl Plants 51) | Coding? Yes |
Transcript features
This table shows how a lncRNA has been classified (high/low confidence) or whether it has been classified as pre-miRNA (go to FAQ for classification criteria). Besides, it displays the lncRNA length and sequence as well as information about the ORF, the coding potential calculated by the Coding Potential Calculator (CPC), and the folding energies calculated by RNAfold.
| Transcript alias / name | Confidence | Pre-miRNA | Length | Orthologous group | Sequence | Details
|
|---|---|---|---|---|---|---|
| Atauschii_AET6Gv20210900.1 AET6Gv20210900.1 |
Low | No | 1363 | OG0000700 | Get ORF Coding potential... Type: noncoding Potential: -1.267 Folding energies... MFEI: -26.764 AMFE: -0.651 GC content %: 41.086 | |
| Atauschii_AET6Gv20210900.11 AET6Gv20210900.11 |
Low | No | 2783 | OG0000700 | Get ORF Coding potential... Type: noncoding Potential: -0.346 Folding energies... MFEI: -32.972 AMFE: -0.665 GC content %: 49.587 | |
| Atauschii_AET6Gv20210900.12 AET6Gv20210900.12 |
Low | No | 2694 | OG0000700 | Get ORF Coding potential... Type: noncoding Potential: -0.161 Folding energies... MFEI: -33.296 AMFE: -0.669 GC content %: 49.777 | |
| Atauschii_AET6Gv20210900.14 AET6Gv20210900.14 |
Low | No | 2322 | OG0000700 | Get ORF Coding potential... Type: noncoding Potential: -0.358 Folding energies... MFEI: -34.065 AMFE: -0.659 GC content %: 51.680 | |
| Atauschii_AET6Gv20210900.15 AET6Gv20210900.15 |
Low | No | 1715 | OG0000700 | Get ORF Coding potential... Type: noncoding Potential: -1.306 Folding energies... MFEI: -28.093 AMFE: -0.674 GC content %: 41.691 | |
| Atauschii_AET6Gv20210900.16 AET6Gv20210900.16 |
Low | No | 2604 | OG0000700 | Get ORF Coding potential... Type: noncoding Potential: -0.501 Folding energies... MFEI: -33.199 AMFE: -0.667 GC content %: 49.770 | |
| Atauschii_AET6Gv20210900.17 AET6Gv20210900.17 |
Low | No | 2669 | OG0000700 | Get ORF Coding potential... Type: noncoding Potential: -0.369 Folding energies... MFEI: -33.099 AMFE: -0.667 GC content %: 49.607 | |
| Atauschii_AET6Gv20210900.18 AET6Gv20210900.18 |
Low | No | 2253 | OG0000700 | Get ORF Coding potential... Type: noncoding Potential: -0.142 Folding energies... MFEI: -34.004 AMFE: -0.662 GC content %: 51.398 | |
| Atauschii_AET6Gv20210900.19 AET6Gv20210900.19 |
Low | No | 2645 | OG0000700 | Get ORF Coding potential... Type: noncoding Potential: -0.349 Folding energies... MFEI: -33.444 AMFE: -0.673 GC content %: 49.716 | |
| Atauschii_AET6Gv20210900.2 AET6Gv20210900.2 |
Low | No | 1777 | OG0000700 | Get ORF Coding potential... Type: noncoding Potential: -0.286 Folding energies... MFEI: -28.599 AMFE: -0.654 GC content %: 43.725 | |
| Atauschii_AET6Gv20210900.20 AET6Gv20210900.20 |
Low | No | 1770 | OG0000700 | Get ORF Coding potential... Type: noncoding Potential: -0.375 Folding energies... MFEI: -28.667 AMFE: -0.649 GC content %: 44.181 | |
| Atauschii_AET6Gv20210900.22 AET6Gv20210900.22 |
Low | No | 2155 | OG0000700 | Get ORF Coding potential... Type: noncoding Potential: -0.341 Folding energies... MFEI: -29.522 AMFE: -0.663 GC content %: 44.548 | |
| Atauschii_AET6Gv20210900.23 AET6Gv20210900.23 |
Low | No | 2153 | OG0000700 | Get ORF Coding potential... Type: noncoding Potential: -0.425 Folding energies... MFEI: -33.832 AMFE: -0.669 GC content %: 50.581 | |
| Atauschii_AET6Gv20210900.24 AET6Gv20210900.24 |
High | No | 441 | OG0000700 | Get ORF Coding potential... Type: noncoding Potential: -1.412 Folding energies... MFEI: -24.036 AMFE: -0.658 GC content %: 36.508 | |
| Atauschii_AET6Gv20210900.5 AET6Gv20210900.5 |
Low | No | 2682 | OG0000700 | Get ORF Coding potential... Type: noncoding Potential: -0.507 Folding energies... MFEI: -32.919 AMFE: -0.664 GC content %: 49.590 | |
| Atauschii_AET6Gv20210900.7 AET6Gv20210900.7 |
Low | No | 1454 | OG0000700 | Get ORF Coding potential... Type: noncoding Potential: -0.115 Folding energies... MFEI: -29.072 AMFE: -0.643 GC content %: 45.186 | |
| Atauschii_AET6Gv20210900.8 AET6Gv20210900.8 |
Low | No | 2782 | OG0000700 | Get ORF Coding potential... Type: noncoding Potential: -0.374 Folding energies... MFEI: -32.861 AMFE: -0.663 GC content %: 49.533 |
Matches to external databases
| Transcript alias | Database | Version | Hit | Evalue |
|---|---|---|---|---|
| Atauschii_AET6Gv20210900.23 | SwissProt | 2021-04-01 | Q9ZU52 | 3.97e-14 |
| Atauschii_AET6Gv20210900.5 | RepBase (RepeatMasker) | 2014-01-31 | DNA/CMC-EnSpm | |
| Atauschii_AET6Gv20210900.18 | SwissProt | 2021-04-01 | Q9ZU52 | 3.4e-18 |
| Atauschii_AET6Gv20210900.18 | RepBase (RepeatMasker) | 2014-01-31 | DNA/CMC-EnSpm | |
| Atauschii_AET6Gv20210900.11 | SwissProt | 2021-04-01 | Q9ZU52 | 5.78e-14 |
| Atauschii_AET6Gv20210900.12 | RepBase (RepeatMasker) | 2014-01-31 | DNA/CMC-EnSpm | |
| Atauschii_AET6Gv20210900.5 | SwissProt | 2021-04-01 | Q9ZU52 | 5.49e-14 |
| Atauschii_AET6Gv20210900.7 | RepBase (RepeatMasker) | 2014-01-31 | DNA/CMC-EnSpm | |
| Atauschii_AET6Gv20210900.19 | SwissProt | 2021-04-01 | Q9ZU52 | 5.39e-14 |
| Atauschii_AET6Gv20210900.19 | RepBase (RepeatMasker) | 2014-01-31 | DNA/CMC-EnSpm |
... further results
Gene models