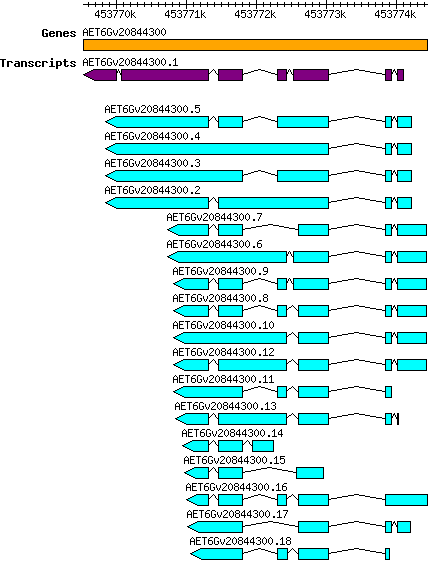
Atauschii AET6Gv20844300
From GreeNC
Gene information
| Gene alias / name | Chrom. | Start | End | Assembly | Species | Details
|
|---|---|---|---|---|---|---|
| Atauschii_AET6Gv20844300 [ AET6Gv20844300] |
6D | 453769532 | 453774433 | v4 | Aegilops tauschii (Ensembl Plants 51) | Coding? Yes |
Transcript features
This table shows how a lncRNA has been classified (high/low confidence) or whether it has been classified as pre-miRNA (go to FAQ for classification criteria). Besides, it displays the lncRNA length and sequence as well as information about the ORF, the coding potential calculated by the Coding Potential Calculator (CPC), and the folding energies calculated by RNAfold.
| Transcript alias / name | Confidence | Pre-miRNA | Length | Orthologous group | Sequence | Details
|
|---|---|---|---|---|---|---|
| Atauschii_AET6Gv20844300.10 AET6Gv20844300.10 |
Low | No | 2564 | OG0000703 | Get ORF Coding potential... Type: noncoding Potential: -1.181 Folding energies... MFEI: -30.445 AMFE: -0.668 GC content %: 45.593 | |
| Atauschii_AET6Gv20844300.11 AET6Gv20844300.11 |
Low | No | 1636 | OG0000703 | Get ORF Coding potential... Type: noncoding Potential: -1.236 Folding energies... MFEI: -27.200 AMFE: -0.615 GC content %: 44.254 | |
| Atauschii_AET6Gv20844300.12 AET6Gv20844300.12 |
Low | No | 2411 | OG0000703 | Get ORF Coding potential... Type: noncoding Potential: -1.175 Folding energies... MFEI: -30.539 AMFE: -0.665 GC content %: 45.915 | |
| Atauschii_AET6Gv20844300.13 AET6Gv20844300.13 |
Low | No | 1989 | OG0000703 | Get ORF Coding potential... Type: noncoding Potential: -1.154 Folding energies... MFEI: -27.049 AMFE: -0.646 GC content %: 41.880 | |
| Atauschii_AET6Gv20844300.14 AET6Gv20844300.14 |
Low | No | 1023 | OG0000703 | Get ORF Coding potential... Type: noncoding Potential: -0.476 Folding energies... MFEI: -25.474 AMFE: -0.612 GC content %: 41.642 | |
| Atauschii_AET6Gv20844300.15 AET6Gv20844300.15 |
Low | No | 1070 | OG0000703 | Get ORF Coding potential... Type: noncoding Potential: -0.484 Folding energies... MFEI: -27.944 AMFE: -0.608 GC content %: 45.981 | |
| Atauschii_AET6Gv20844300.16 AET6Gv20844300.16 |
Low | No | 1819 | OG0000703 | Get ORF Coding potential... Type: noncoding Potential: -1.216 Folding energies... MFEI: -33.535 AMFE: -0.651 GC content %: 51.512 | |
| Atauschii_AET6Gv20844300.17 AET6Gv20844300.17 |
Low | No | 1493 | OG0000703 | Get ORF Coding potential... Type: noncoding Potential: -1.227 Folding energies... MFEI: -30.884 AMFE: -0.640 GC content %: 48.292 | |
| Atauschii_AET6Gv20844300.18 AET6Gv20844300.18 |
Low | No | 1388 | OG0000703 | Get ORF Coding potential... Type: noncoding Potential: -1.219 Folding energies... MFEI: -28.869 AMFE: -0.626 GC content %: 46.110 | |
| Atauschii_AET6Gv20844300.2 AET6Gv20844300.2 |
Low | No | 3325 | OG0000703 | Get ORF Coding potential... Type: noncoding Potential: -1.203 Folding energies... MFEI: -29.239 AMFE: -0.689 GC content %: 42.436 | |
| Atauschii_AET6Gv20844300.3 AET6Gv20844300.3 |
Low | No | 2956 | OG0000703 | Get ORF Coding potential... Type: noncoding Potential: -1.279 Folding energies... MFEI: -29.648 AMFE: -0.674 GC content %: 43.978 | |
| Atauschii_AET6Gv20844300.4 AET6Gv20844300.4 |
Low | No | 3464 | OG0000703 | Get ORF Coding potential... Type: noncoding Potential: -1.206 Folding energies... MFEI: -29.111 AMFE: -0.686 GC content %: 42.408 | |
| Atauschii_AET6Gv20844300.5 AET6Gv20844300.5 |
Low | No | 2817 | OG0000703 | Get ORF Coding potential... Type: noncoding Potential: -1.239 Folding energies... MFEI: -29.826 AMFE: -0.676 GC content %: 44.089 | |
| Atauschii_AET6Gv20844300.6 AET6Gv20844300.6 |
Low | No | 2715 | OG0000703 | Get ORF Coding potential... Type: noncoding Potential: -1.187 Folding energies... MFEI: -30.243 AMFE: -0.675 GC content %: 44.825 | |
| Atauschii_AET6Gv20844300.7 AET6Gv20844300.7 |
Low | No | 1879 | OG0000703 | Get ORF Coding potential... Type: noncoding Potential: -1.218 Folding energies... MFEI: -31.879 AMFE: -0.655 GC content %: 48.696 | |
| Atauschii_AET6Gv20844300.8 AET6Gv20844300.8 |
Low | No | 1917 | OG0000703 | Get ORF Coding potential... Type: noncoding Potential: -1.219 Folding energies... MFEI: -31.805 AMFE: -0.647 GC content %: 49.139 | |
| Atauschii_AET6Gv20844300.9 AET6Gv20844300.9 |
Low | No | 1979 | OG0000703 | Get ORF Coding potential... Type: noncoding Potential: -1.221 Folding energies... MFEI: -31.890 AMFE: -0.655 GC content %: 48.711 |
Matches to external databases
| Transcript alias | Database | Version | Hit | Evalue |
|---|---|---|---|---|
| Atauschii_AET6Gv20844300.10 | SwissProt | 2021-04-01 | Q02764 | 2.81e-5 |
| Atauschii_AET6Gv20844300.9 | SwissProt | 2021-04-01 | Q02764 | 2.17e-10 |
| Atauschii_AET6Gv20844300.4 | SwissProt | 2021-04-01 | Q02764 | 3.94e-5 |
| Atauschii_AET6Gv20844300.16 | SwissProt | 2021-04-01 | Q02764 | 1.95e-10 |
| Atauschii_AET6Gv20844300.8 | RepBase (RepeatMasker) | 2014-01-31 | LINE/L1 | |
| Atauschii_AET6Gv20844300.11 | SwissProt | 2021-04-01 | Q02764 | 1.65e-5 |
| Atauschii_AET6Gv20844300.10 | RepBase (RepeatMasker) | 2014-01-31 | LINE/L1 | |
| Atauschii_AET6Gv20844300.5 | SwissProt | 2021-04-01 | Q02764 | 3.28e-10 |
| Atauschii_AET6Gv20844300.17 | SwissProt | 2021-04-01 | Q02764 | 1.47e-5 |
| Atauschii_AET6Gv20844300.12 | SwissProt | 2021-04-01 | Q02764 | 2.74e-10 |
... further results
Gene models