Atauschii AET7Gv20759100
From GreeNC
Gene information
| Gene alias / name | Chrom. | Start | End | Assembly | Species | Details
|
|---|---|---|---|---|---|---|
| Atauschii_AET7Gv20759100 [ AET7Gv20759100] |
7D | 378555455 | 378561021 | v4 | Aegilops tauschii (Ensembl Plants 51) | Coding? No |
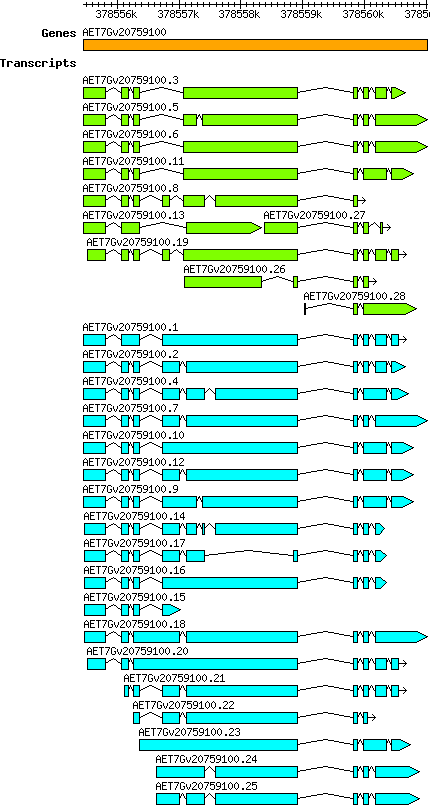
Transcript features
This table shows how a lncRNA has been classified (high/low confidence) or whether it has been classified as pre-miRNA (go to FAQ for classification criteria). Besides, it displays the lncRNA length and sequence as well as information about the ORF, the coding potential calculated by the Coding Potential Calculator (CPC), and the folding energies calculated by RNAfold.
| Transcript alias / name | Confidence | Pre-miRNA | Length | Orthologous group | Sequence | Details
|
|---|---|---|---|---|---|---|
| Atauschii_AET7Gv20759100.1 AET7Gv20759100.1 |
Low | No | 3284 | OG0000228 | Get ORF Coding potential... Type: noncoding Potential: -1.020 Folding energies... MFEI: -29.083 AMFE: -0.670 GC content %: 43.392 | |
| Atauschii_AET7Gv20759100.10 AET7Gv20759100.10 |
Low | No | 3546 | OG0000228 | Get ORF Coding potential... Type: noncoding Potential: -0.893 Folding energies... MFEI: -29.013 AMFE: -0.673 GC content %: 43.091 | |
| Atauschii_AET7Gv20759100.11 AET7Gv20759100.11 |
High | No | 3200 | OG0000228 | Get ORF Coding potential... Type: noncoding Potential: -0.883 Folding energies... MFEI: -29.322 AMFE: -0.676 GC content %: 43.406 | |
| Atauschii_AET7Gv20759100.12 AET7Gv20759100.12 |
Low | No | 3426 | OG0000228 | Get ORF Coding potential... Type: noncoding Potential: -0.889 Folding energies... MFEI: -29.104 AMFE: -0.676 GC content %: 43.082 | |
| Atauschii_AET7Gv20759100.13 AET7Gv20759100.13 |
High | No | 1868 | OG0000228 | Get ORF Coding potential... Type: noncoding Potential: -0.946 Folding energies... MFEI: -30.696 AMFE: -0.669 GC content %: 45.878 | |
| Atauschii_AET7Gv20759100.14 AET7Gv20759100.14 |
Low | No | 2651 | OG0000228 | Get ORF Coding potential... Type: noncoding Potential: -0.634 Folding energies... MFEI: -29.374 AMFE: -0.665 GC content %: 44.172 | |
| Atauschii_AET7Gv20759100.15 AET7Gv20759100.15 |
Low | No | 848 | OG0000228 | Get ORF Coding potential... Type: noncoding Potential: -0.772 Folding energies... MFEI: -33.373 AMFE: -0.633 GC content %: 52.712 | |
| Atauschii_AET7Gv20759100.16 AET7Gv20759100.16 |
Low | No | 3070 | OG0000228 | Get ORF Coding potential... Type: noncoding Potential: -1.013 Folding energies... MFEI: -28.785 AMFE: -0.665 GC content %: 43.290 | |
| Atauschii_AET7Gv20759100.17 AET7Gv20759100.17 |
Low | No | 1510 | OG0000228 | Get ORF Coding potential... Type: noncoding Potential: -0.908 Folding energies... MFEI: -31.338 AMFE: -0.624 GC content %: 50.199 | |
| Atauschii_AET7Gv20759100.18 AET7Gv20759100.18 |
Low | No | 3984 | OG0000228 | Get ORF Coding potential... Type: noncoding Potential: -1.007 Folding energies... MFEI: -28.341 AMFE: -0.681 GC content %: 41.616 | |
| Atauschii_AET7Gv20759100.19 AET7Gv20759100.19 |
High | No | 2903 | OG0000228 | Get ORF Coding potential... Type: noncoding Potential: -1.001 Folding energies... MFEI: -28.980 AMFE: -0.671 GC content %: 43.162 | |
| Atauschii_AET7Gv20759100.2 AET7Gv20759100.2 |
Low | No | 3176 | OG0000228 | Get ORF Coding potential... Type: noncoding Potential: -1.017 Folding energies... MFEI: -29.364 AMFE: -0.675 GC content %: 43.514 | |
| Atauschii_AET7Gv20759100.20 AET7Gv20759100.20 |
Low | No | 3502 | OG0000228 | Get ORF Coding potential... Type: noncoding Potential: -1.019 Folding energies... MFEI: -28.718 AMFE: -0.683 GC content %: 42.062 | |
| Atauschii_AET7Gv20759100.21 AET7Gv20759100.21 |
Low | No | 2664 | OG0000228 | Get ORF Coding potential... Type: noncoding Potential: -1.032 Folding energies... MFEI: -27.399 AMFE: -0.670 GC content %: 40.916 | |
| Atauschii_AET7Gv20759100.22 AET7Gv20759100.22 |
Low | No | 2287 | OG0000228 | Get ORF Coding potential... Type: noncoding Potential: -1.025 Folding energies... MFEI: -26.366 AMFE: -0.661 GC content %: 39.878 | |
| Atauschii_AET7Gv20759100.23 AET7Gv20759100.23 |
Low | No | 3296 | OG0000228 | Get ORF Coding potential... Type: noncoding Potential: -0.886 Folding energies... MFEI: -26.584 AMFE: -0.676 GC content %: 39.351 | |
| Atauschii_AET7Gv20759100.24 AET7Gv20759100.24 |
Low | No | 2965 | OG0000228 | Get ORF Coding potential... Type: noncoding Potential: -0.984 Folding energies... MFEI: -26.671 AMFE: -0.668 GC content %: 39.899 | |
| Atauschii_AET7Gv20759100.25 AET7Gv20759100.25 |
Low | No | 2845 | OG0000228 | Get ORF Coding potential... Type: noncoding Potential: -0.981 Folding energies... MFEI: -26.731 AMFE: -0.672 GC content %: 39.754 | |
| Atauschii_AET7Gv20759100.26 AET7Gv20759100.26 |
High | No | 1476 | OG0000228 | Get ORF Coding potential... Type: noncoding Potential: -1.154 Folding energies... MFEI: -27.121 AMFE: -0.668 GC content %: 40.583 | |
| Atauschii_AET7Gv20759100.27 AET7Gv20759100.27 |
High | No | 713 | OG0000228 | Get ORF Coding potential... Type: noncoding Potential: -1.066 Folding energies... MFEI: -23.506 AMFE: -0.605 GC content %: 38.850 | |
| Atauschii_AET7Gv20759100.28 AET7Gv20759100.28 |
High | No | 953 | OG0000228 | Get ORF Coding potential... Type: noncoding Potential: -0.820 Folding energies... MFEI: -26.684 AMFE: -0.647 GC content %: 41.238 | |
| Atauschii_AET7Gv20759100.3 AET7Gv20759100.3 |
High | No | 2950 | OG0000228 | Get ORF Coding potential... Type: noncoding Potential: -1.009 Folding energies... MFEI: -29.624 AMFE: -0.675 GC content %: 43.898 | |
| Atauschii_AET7Gv20759100.4 AET7Gv20759100.4 |
Low | No | 3166 | OG0000228 | Get ORF Coding potential... Type: noncoding Potential: -0.837 Folding energies... MFEI: -29.431 AMFE: -0.670 GC content %: 43.904 | |
| Atauschii_AET7Gv20759100.5 AET7Gv20759100.5 |
High | No | 3298 | OG0000228 | Get ORF Coding potential... Type: noncoding Potential: -0.970 Folding energies... MFEI: -29.175 AMFE: -0.684 GC content %: 42.662 | |
| Atauschii_AET7Gv20759100.6 AET7Gv20759100.6 |
High | No | 3395 | OG0000228 | Get ORF Coding potential... Type: noncoding Potential: -0.995 Folding energies... MFEI: -28.996 AMFE: -0.679 GC content %: 42.680 | |
| Atauschii_AET7Gv20759100.7 AET7Gv20759100.7 |
Low | No | 3621 | OG0000228 | Get ORF Coding potential... Type: noncoding Potential: -1.000 Folding energies... MFEI: -28.810 AMFE: -0.679 GC content %: 42.419 | |
| Atauschii_AET7Gv20759100.8 AET7Gv20759100.8 |
High | No | 2415 | OG0000228 | Get ORF Coding potential... Type: noncoding Potential: -0.984 Folding energies... MFEI: -29.416 AMFE: -0.668 GC content %: 44.058 | |
| Atauschii_AET7Gv20759100.9 AET7Gv20759100.9 |
Low | No | 3448 | OG0000228 | Get ORF Coding potential... Type: noncoding Potential: -0.890 Folding energies... MFEI: -29.115 AMFE: -0.676 GC content %: 43.097 |
Matches to external databases
| Transcript alias | Database | Version | Hit | Evalue |
|---|---|---|---|---|
| Atauschii_AET7Gv20759100.17 | SwissProt | 2021-04-01 | F4JG10 | 3.03e-6 |
| Atauschii_AET7Gv20759100.10 | SwissProt | 2021-04-01 | A0A1P8 | 2.67e-5 |
| Atauschii_AET7Gv20759100.9 | SwissProt | 2021-04-01 | A0A1P8 | 2.59e-5 |
| Atauschii_AET7Gv20759100.23 | SwissProt | 2021-04-01 | A0A1P8 | 1.57e-5 |
| Atauschii_AET7Gv20759100.18 | SwissProt | 2021-04-01 | F4JG10 | 1.25e-5 |
| Atauschii_AET7Gv20759100.12 | SwissProt | 2021-04-01 | F4JG10 | 1.67e-5 |
| Atauschii_AET7Gv20759100.24 | SwissProt | 2021-04-01 | O80596 | 8.77e-6 |
| Atauschii_AET7Gv20759100.2 | SwissProt | 2021-04-01 | F4JG10 | 1.54e-5 |
| Atauschii_AET7Gv20759100.14 | SwissProt | 2021-04-01 | F4JG10 | 1.27e-5 |
| Atauschii_AET7Gv20759100.25 | SwissProt | 2021-04-01 | O80596 | 1.38e-6 |
... further results
Gene models