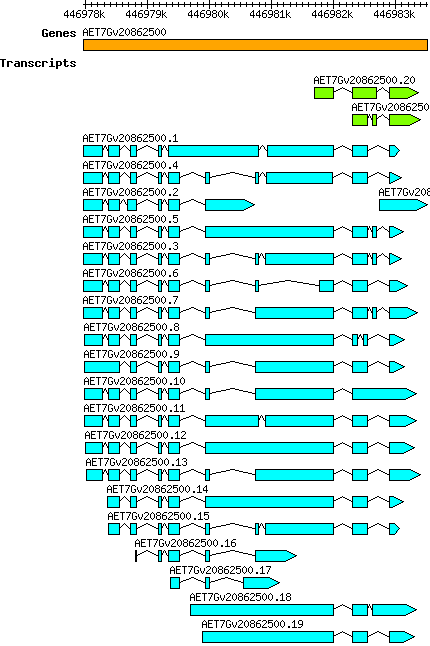
Atauschii AET7Gv20862500
From GreeNC
Gene information
| Gene alias / name | Chrom. | Start | End | Assembly | Species | Details
|
|---|---|---|---|---|---|---|
| Atauschii_AET7Gv20862500 [ AET7Gv20862500] |
7D | 446977971 | 446983510 | v4 | Aegilops tauschii (Ensembl Plants 51) | Coding? No |
Transcript features
This table shows how a lncRNA has been classified (high/low confidence) or whether it has been classified as pre-miRNA (go to FAQ for classification criteria). Besides, it displays the lncRNA length and sequence as well as information about the ORF, the coding potential calculated by the Coding Potential Calculator (CPC), and the folding energies calculated by RNAfold.
| Transcript alias / name | Confidence | Pre-miRNA | Length | Orthologous group | Sequence | Details
|
|---|---|---|---|---|---|---|
| Atauschii_AET7Gv20862500.1 AET7Gv20862500.1 |
Low | No | 3525 | OG0000406 | Get ORF Coding potential... Type: noncoding Potential: -0.929 Folding energies... MFEI: -28.664 AMFE: -0.662 GC content %: 43.291 | |
| Atauschii_AET7Gv20862500.10 AET7Gv20862500.10 |
Low | No | 3137 | OG0000406 | Get ORF Coding potential... Type: noncoding Potential: -0.197 Folding energies... MFEI: -28.948 AMFE: -0.642 GC content %: 45.107 | |
| Atauschii_AET7Gv20862500.11 AET7Gv20862500.11 |
Low | No | 3403 | OG0000406 | Get ORF Coding potential... Type: noncoding Potential: -0.207 Folding energies... MFEI: -28.813 AMFE: -0.657 GC content %: 43.873 | |
| Atauschii_AET7Gv20862500.12 AET7Gv20862500.12 |
Low | No | 3464 | OG0000406 | Get ORF Coding potential... Type: noncoding Potential: -0.210 Folding energies... MFEI: -28.568 AMFE: -0.661 GC content %: 43.245 | |
| Atauschii_AET7Gv20862500.13 AET7Gv20862500.13 |
Low | No | 2816 | OG0000406 | Get ORF Coding potential... Type: noncoding Potential: -0.189 Folding energies... MFEI: -30.107 AMFE: -0.659 GC content %: 45.668 | |
| Atauschii_AET7Gv20862500.14 AET7Gv20862500.14 |
Low | No | 3043 | OG0000406 | Get ORF Coding potential... Type: noncoding Potential: -0.199 Folding energies... MFEI: -27.805 AMFE: -0.665 GC content %: 41.834 | |
| Atauschii_AET7Gv20862500.15 AET7Gv20862500.15 |
Low | No | 2113 | OG0000406 | Get ORF Coding potential... Type: noncoding Potential: -0.160 Folding energies... MFEI: -29.584 AMFE: -0.653 GC content %: 45.338 | |
| Atauschii_AET7Gv20862500.16 AET7Gv20862500.16 |
Low | No | 967 | OG0000406 | Get ORF Coding potential... Type: noncoding Potential: -0.277 Folding energies... MFEI: -28.325 AMFE: -0.657 GC content %: 43.123 | |
| Atauschii_AET7Gv20862500.17 AET7Gv20862500.17 |
Low | No | 789 | OG0000406 | Get ORF Coding potential... Type: noncoding Potential: -0.094 Folding energies... MFEI: -26.768 AMFE: -0.640 GC content %: 41.825 | |
| Atauschii_AET7Gv20862500.18 AET7Gv20862500.18 |
Low | No | 3237 | OG0000406 | Get ORF Coding potential... Type: noncoding Potential: -0.828 Folding energies... MFEI: -24.445 AMFE: -0.633 GC content %: 38.616 | |
| Atauschii_AET7Gv20862500.19 AET7Gv20862500.19 |
Low | No | 2738 | OG0000406 | Get ORF Coding potential... Type: noncoding Potential: -0.998 Folding energies... MFEI: -24.748 AMFE: -0.636 GC content %: 38.934 | |
| Atauschii_AET7Gv20862500.2 AET7Gv20862500.2 |
Low | No | 1644 | OG0000406 | Get ORF Coding potential... Type: noncoding Potential: -0.178 Folding energies... MFEI: -30.645 AMFE: -0.663 GC content %: 46.229 | |
| Atauschii_AET7Gv20862500.20 AET7Gv20862500.20 |
High | No | 1134 | OG0000406 | Get ORF Coding potential... Type: noncoding Potential: -0.904 Folding energies... MFEI: -23.623 AMFE: -0.539 GC content %: 43.827 | |
| Atauschii_AET7Gv20862500.21 AET7Gv20862500.21 |
High | No | 803 | OG0000406 | Get ORF Coding potential... Type: noncoding Potential: -0.683 Folding energies... MFEI: -23.898 AMFE: -0.564 GC content %: 42.341 | |
| Atauschii_AET7Gv20862500.22 AET7Gv20862500.22 |
Low | Yes | 770 | OG0000406 | Get ORF Coding potential... Type: noncoding Potential: -1.380 Folding energies... MFEI: -23.312 AMFE: -0.606 GC content %: 38.442 | |
| Atauschii_AET7Gv20862500.3 AET7Gv20862500.3 |
Low | No | 2496 | OG0000406 | Get ORF Coding potential... Type: noncoding Potential: -0.092 Folding energies... MFEI: -30.717 AMFE: -0.646 GC content %: 47.516 | |
| Atauschii_AET7Gv20862500.4 AET7Gv20862500.4 |
Low | No | 2404 | OG0000406 | Get ORF Coding potential... Type: noncoding Potential: -0.178 Folding energies... MFEI: -30.874 AMFE: -0.647 GC content %: 47.712 | |
| Atauschii_AET7Gv20862500.5 AET7Gv20862500.5 |
Low | No | 3387 | OG0000406 | Get ORF Coding potential... Type: noncoding Potential: -0.125 Folding energies... MFEI: -28.636 AMFE: -0.654 GC content %: 43.785 | |
| Atauschii_AET7Gv20862500.6 AET7Gv20862500.6 |
Low | No | 1654 | OG0000406 | Get ORF Coding potential... Type: noncoding Potential: -0.157 Folding energies... MFEI: -32.304 AMFE: -0.627 GC content %: 51.511 | |
| Atauschii_AET7Gv20862500.7 AET7Gv20862500.7 |
Low | No | 2865 | OG0000406 | Get ORF Coding potential... Type: noncoding Potential: -0.105 Folding energies... MFEI: -29.815 AMFE: -0.645 GC content %: 46.248 | |
| Atauschii_AET7Gv20862500.8 AET7Gv20862500.8 |
Low | No | 3240 | OG0000406 | Get ORF Coding potential... Type: noncoding Potential: -0.205 Folding energies... MFEI: -28.793 AMFE: -0.662 GC content %: 43.519 | |
| Atauschii_AET7Gv20862500.9 AET7Gv20862500.9 |
Low | No | 2702 | OG0000406 | Get ORF Coding potential... Type: noncoding Potential: -0.183 Folding energies... MFEI: -30.163 AMFE: -0.652 GC content %: 46.262 |
Matches to external databases
| Transcript alias | Database | Version | Hit | Evalue |
|---|---|---|---|---|
| Atauschii_AET7Gv20862500.12 | SwissProt | 2021-04-01 | Q9ZV71 | 3.86e-12 |
| Atauschii_AET7Gv20862500.22 | Rfam | 14.6 | MIR1122 | 8.3e-7 |
| Atauschii_AET7Gv20862500.9 | RepBase (RepeatMasker) | 2014-01-31 | LTR/Copia | |
| Atauschii_AET7Gv20862500.5 | SwissProt | 2021-04-01 | Q9ZV71 | 3.76e-12 |
| Atauschii_AET7Gv20862500.3 | RepBase (RepeatMasker) | 2014-01-31 | LTR/Copia | |
| Atauschii_AET7Gv20862500.18 | SwissProt | 2021-04-01 | Q9LZD3 | 6.14e-10 |
| Atauschii_AET7Gv20862500.14 | RepBase (RepeatMasker) | 2014-01-31 | LTR/Copia | |
| Atauschii_AET7Gv20862500.13 | SwissProt | 2021-04-01 | Q9ZV71 | 3.09e-12 |
| Atauschii_AET7Gv20862500.1 | RepBase (RepeatMasker) | 2014-01-31 | LTR/Copia | |
| Atauschii_AET7Gv20862500.6 | SwissProt | 2021-04-01 | Q9ZV71 | 1.68e-12 |
... further results
Gene models