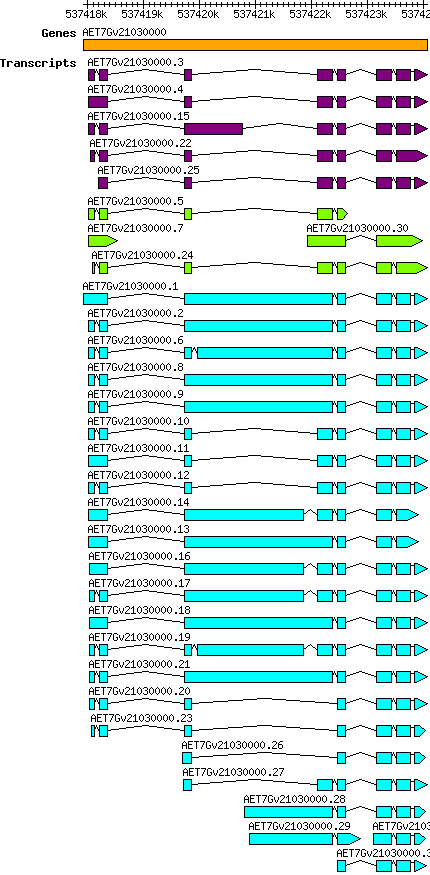
Atauschii AET7Gv21030000
From GreeNC
Gene information
| Gene alias / name | Chrom. | Start | End | Assembly | Species | Details
|
|---|---|---|---|---|---|---|
| Atauschii_AET7Gv21030000 [ AET7Gv21030000] |
7D | 537417926 | 537424077 | v4 | Aegilops tauschii (Ensembl Plants 51) | Coding? Yes |
Transcript features
This table shows how a lncRNA has been classified (high/low confidence) or whether it has been classified as pre-miRNA (go to FAQ for classification criteria). Besides, it displays the lncRNA length and sequence as well as information about the ORF, the coding potential calculated by the Coding Potential Calculator (CPC), and the folding energies calculated by RNAfold.
| Transcript alias / name | Confidence | Pre-miRNA | Length | Orthologous group | Sequence | Details
|
|---|---|---|---|---|---|---|
| Atauschii_AET7Gv21030000.1 AET7Gv21030000.1 |
Low | No | 3957 | OG0000091 | Get ORF Coding potential... Type: noncoding Potential: -1.000 Folding energies... MFEI: -33.192 AMFE: -0.668 GC content %: 49.684 | |
| Atauschii_AET7Gv21030000.10 AET7Gv21030000.10 |
Low | No | 1536 | OG0000091 | Get ORF Coding potential... Type: noncoding Potential: -0.931 Folding energies... MFEI: -33.900 AMFE: -0.640 GC content %: 52.930 | |
| Atauschii_AET7Gv21030000.11 AET7Gv21030000.11 |
Low | No | 1615 | OG0000091 | Get ORF Coding potential... Type: noncoding Potential: -0.936 Folding energies... MFEI: -33.505 AMFE: -0.641 GC content %: 52.260 | |
| Atauschii_AET7Gv21030000.12 AET7Gv21030000.12 |
Low | No | 1529 | OG0000091 | Get ORF Coding potential... Type: noncoding Potential: -0.931 Folding energies... MFEI: -33.996 AMFE: -0.643 GC content %: 52.845 | |
| Atauschii_AET7Gv21030000.13 AET7Gv21030000.13 |
Low | No | 3771 | OG0000091 | Get ORF Coding potential... Type: noncoding Potential: -1.141 Folding energies... MFEI: -32.798 AMFE: -0.672 GC content %: 48.820 | |
| Atauschii_AET7Gv21030000.14 AET7Gv21030000.14 |
Low | No | 3524 | OG0000091 | Get ORF Coding potential... Type: noncoding Potential: -1.139 Folding energies... MFEI: -33.371 AMFE: -0.670 GC content %: 49.773 | |
| Atauschii_AET7Gv21030000.16 AET7Gv21030000.16 |
Low | No | 3608 | OG0000091 | Get ORF Coding potential... Type: noncoding Potential: -0.996 Folding energies... MFEI: -33.700 AMFE: -0.673 GC content %: 50.083 | |
| Atauschii_AET7Gv21030000.17 AET7Gv21030000.17 |
Low | No | 3522 | OG0000091 | Get ORF Coding potential... Type: noncoding Potential: -0.994 Folding energies... MFEI: -33.938 AMFE: -0.675 GC content %: 50.312 | |
| Atauschii_AET7Gv21030000.18 AET7Gv21030000.18 |
Low | No | 3853 | OG0000091 | Get ORF Coding potential... Type: noncoding Potential: -0.999 Folding energies... MFEI: -33.156 AMFE: -0.674 GC content %: 49.157 | |
| Atauschii_AET7Gv21030000.19 AET7Gv21030000.19 |
Low | No | 3414 | OG0000091 | Get ORF Coding potential... Type: noncoding Potential: -0.993 Folding energies... MFEI: -34.171 AMFE: -0.674 GC content %: 50.732 | |
| Atauschii_AET7Gv21030000.2 AET7Gv21030000.2 |
Low | No | 3785 | OG0000091 | Get ORF Coding potential... Type: noncoding Potential: -0.998 Folding energies... MFEI: -33.186 AMFE: -0.672 GC content %: 49.353 | |
| Atauschii_AET7Gv21030000.20 AET7Gv21030000.20 |
Low | No | 1243 | OG0000091 | Get ORF Coding potential... Type: noncoding Potential: -0.908 Folding energies... MFEI: -35.390 AMFE: -0.650 GC content %: 54.465 | |
| Atauschii_AET7Gv21030000.21 AET7Gv21030000.21 |
Low | No | 3754 | OG0000091 | Get ORF Coding potential... Type: noncoding Potential: -0.998 Folding energies... MFEI: -33.418 AMFE: -0.678 GC content %: 49.281 | |
| Atauschii_AET7Gv21030000.23 AET7Gv21030000.23 |
Low | No | 1177 | OG0000091 | Get ORF Coding potential... Type: noncoding Potential: -0.902 Folding energies... MFEI: -33.968 AMFE: -0.629 GC content %: 54.036 | |
| Atauschii_AET7Gv21030000.24 AET7Gv21030000.24 |
High | No | 1530 | OG0000091 | Get ORF Coding potential... Type: noncoding Potential: -1.131 Folding energies... MFEI: -33.052 AMFE: -0.644 GC content %: 51.307 | |
| Atauschii_AET7Gv21030000.26 AET7Gv21030000.26 |
Low | No | 997 | OG0000091 | Get ORF Coding potential... Type: noncoding Potential: -0.882 Folding energies... MFEI: -31.966 AMFE: -0.618 GC content %: 51.755 | |
| Atauschii_AET7Gv21030000.27 AET7Gv21030000.27 |
Low | No | 1293 | OG0000091 | Get ORF Coding potential... Type: noncoding Potential: -0.913 Folding energies... MFEI: -31.454 AMFE: -0.625 GC content %: 50.348 | |
| Atauschii_AET7Gv21030000.28 AET7Gv21030000.28 |
Low | No | 2407 | OG0000091 | Get ORF Coding potential... Type: noncoding Potential: -0.971 Folding energies... MFEI: -28.903 AMFE: -0.659 GC content %: 43.830 | |
| Atauschii_AET7Gv21030000.29 AET7Gv21030000.29 |
Low | No | 1876 | OG0000091 | Get ORF Coding potential... Type: noncoding Potential: -1.376 Folding energies... MFEI: -26.205 AMFE: -0.638 GC content %: 41.045 | |
| Atauschii_AET7Gv21030000.30 AET7Gv21030000.30 |
High | No | 1507 | OG0000091 | Get ORF Coding potential... Type: noncoding Potential: -1.092 Folding energies... MFEI: -29.051 AMFE: -0.643 GC content %: 45.189 | |
| Atauschii_AET7Gv21030000.31 AET7Gv21030000.31 |
Low | No | 873 | OG0000091 | Get ORF Coding potential... Type: noncoding Potential: -0.865 Folding energies... MFEI: -30.825 AMFE: -0.610 GC content %: 50.515 | |
| Atauschii_AET7Gv21030000.32 AET7Gv21030000.32 |
Low | No | 760 | OG0000091 | Get ORF Coding potential... Type: noncoding Potential: -0.847 Folding energies... MFEI: -30.539 AMFE: -0.627 GC content %: 48.684 | |
| Atauschii_AET7Gv21030000.5 AET7Gv21030000.5 |
High | No | 824 | OG0000091 | Get ORF Coding potential... Type: noncoding Potential: -1.075 Folding energies... MFEI: -35.085 AMFE: -0.634 GC content %: 55.340 | |
| Atauschii_AET7Gv21030000.6 AET7Gv21030000.6 |
Low | No | 3675 | OG0000091 | Get ORF Coding potential... Type: noncoding Potential: -0.997 Folding energies... MFEI: -33.445 AMFE: -0.673 GC content %: 49.687 | |
| Atauschii_AET7Gv21030000.7 AET7Gv21030000.7 |
High | No | 513 | OG0081374 | Get ORF Coding potential... Type: noncoding Potential: -1.007 Folding energies... MFEI: -29.961 AMFE: -0.571 GC content %: 52.437 | |
| Atauschii_AET7Gv21030000.8 AET7Gv21030000.8 |
Low | No | 3787 | OG0000091 | Get ORF Coding potential... Type: noncoding Potential: -0.998 Folding energies... MFEI: -33.214 AMFE: -0.673 GC content %: 49.379 | |
| Atauschii_AET7Gv21030000.9 AET7Gv21030000.9 |
Low | No | 3783 | OG0000091 | Get ORF Coding potential... Type: noncoding Potential: -0.998 Folding energies... MFEI: -33.238 AMFE: -0.673 GC content %: 49.379 |
Matches to external databases
| Transcript alias | Database | Version | Hit | Evalue |
|---|---|---|---|---|
| Atauschii_AET7Gv21030000.9 | RepBase (RepeatMasker) | 2014-01-31 | DNA/TcMar-Mariner; DNA/TcMar-Stowaway | |
| Atauschii_AET7Gv21030000.10 | SwissProt | 2021-04-01 | Q9ZVC9 | 2.15e-6 |
| Atauschii_AET7Gv21030000.6 | SwissProt | 2021-04-01 | Q9ZVC9 | 7.24e-6 |
| Atauschii_AET7Gv21030000.26 | SwissProt | 2021-04-01 | Q9ZVC9 | 8.9e-8 |
| Atauschii_AET7Gv21030000.21 | RepBase (RepeatMasker) | 2014-01-31 | DNA/TcMar-Mariner; DNA/TcMar-Stowaway | |
| Atauschii_AET7Gv21030000.19 | SwissProt | 2021-04-01 | Q9ZVC9 | 6.62e-6 |
| Atauschii_AET7Gv21030000.16 | RepBase (RepeatMasker) | 2014-01-31 | DNA/TcMar-Mariner; DNA/TcMar-Stowaway | |
| Atauschii_AET7Gv21030000.11 | SwissProt | 2021-04-01 | Q9ZVC9 | 2.33e-6 |
| Atauschii_AET7Gv21030000.8 | SwissProt | 2021-04-01 | Q9ZVC9 | 7.51e-6 |
| Atauschii_AET7Gv21030000.27 | SwissProt | 2021-04-01 | Q9ZVC9 | 1.56e-6 |
... further results
Gene models