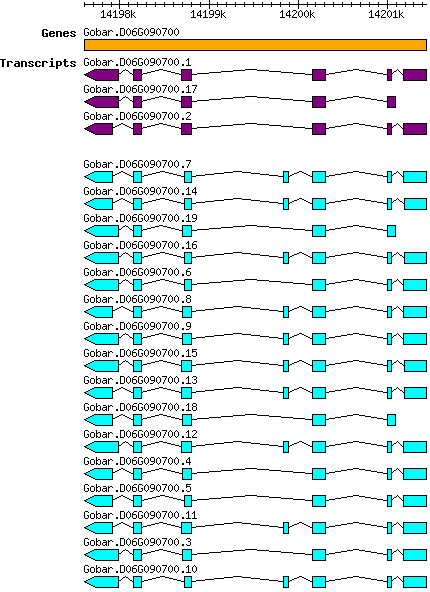
Gbarbadense Gobar.D06G090700
From GreeNC
Gene information
| Gene alias / name | Chrom. | Start | End | Assembly | Species | Details
|
|---|---|---|---|---|---|---|
| Gbarbadense_Gobar.D06G090700 [ Gobar.D06G090700] |
D06 | 14197607 | 14201435 | v1.1 | Gossypium barbadense (Phytozome 13) | Coding? Yes |
Transcript features
This table shows how a lncRNA has been classified (high/low confidence) or whether it has been classified as pre-miRNA (go to FAQ for classification criteria). Besides, it displays the lncRNA length and sequence as well as information about the ORF, the coding potential calculated by the Coding Potential Calculator (CPC), and the folding energies calculated by RNAfold.
| Transcript alias / name | Confidence | Pre-miRNA | Length | Orthologous group | Sequence | Details
|
|---|---|---|---|---|---|---|
| Gbarbadense_Gobar.D06G090700.10 Gobar.D06G090700.10 |
Low | No | 1052 | OG0000094 | Get ORF Coding potential... Type: noncoding Potential: -0.808 Folding energies... MFEI: -23.983 AMFE: -0.615 GC content %: 38.973 | |
| Gbarbadense_Gobar.D06G090700.11 Gobar.D06G090700.11 |
Low | No | 1034 | OG0000094 | Get ORF Coding potential... Type: noncoding Potential: -0.804 Folding energies... MFEI: -23.211 AMFE: -0.597 GC content %: 38.878 | |
| Gbarbadense_Gobar.D06G090700.12 Gobar.D06G090700.12 |
Low | No | 1095 | OG0000094 | Get ORF Coding potential... Type: noncoding Potential: -0.816 Folding energies... MFEI: -24.046 AMFE: -0.615 GC content %: 39.087 | |
| Gbarbadense_Gobar.D06G090700.13 Gobar.D06G090700.13 |
Low | No | 1017 | OG0000094 | Get ORF Coding potential... Type: noncoding Potential: -0.543 Folding energies... MFEI: -23.068 AMFE: -0.589 GC content %: 39.135 | |
| Gbarbadense_Gobar.D06G090700.14 Gobar.D06G090700.14 |
Low | No | 974 | OG0000094 | Get ORF Coding potential... Type: noncoding Potential: -0.538 Folding energies... MFEI: -22.659 AMFE: -0.581 GC content %: 39.014 | |
| Gbarbadense_Gobar.D06G090700.15 Gobar.D06G090700.15 |
Low | No | 1078 | OG0000094 | Get ORF Coding potential... Type: noncoding Potential: -0.549 Folding energies... MFEI: -23.924 AMFE: -0.608 GC content %: 39.332 | |
| Gbarbadense_Gobar.D06G090700.16 Gobar.D06G090700.16 |
Low | No | 1035 | OG0000094 | Get ORF Coding potential... Type: noncoding Potential: -0.545 Folding energies... MFEI: -23.855 AMFE: -0.608 GC content %: 39.227 | |
| Gbarbadense_Gobar.D06G090700.18 Gobar.D06G090700.18 |
Low | No | 744 | OG0000094 | Get ORF Coding potential... Type: noncoding Potential: -0.933 Folding energies... MFEI: -24.866 AMFE: -0.685 GC content %: 36.290 | |
| Gbarbadense_Gobar.D06G090700.19 Gobar.D06G090700.19 |
Low | No | 805 | OG0000094 | Get ORF Coding potential... Type: noncoding Potential: -0.953 Folding energies... MFEI: -24.981 AMFE: -0.679 GC content %: 36.770 | |
| Gbarbadense_Gobar.D06G090700.3 Gobar.D06G090700.3 |
Low | No | 1017 | OG0000094 | Get ORF Coding potential... Type: noncoding Potential: -0.937 Folding energies... MFEI: -23.245 AMFE: -0.600 GC content %: 38.741 | |
| Gbarbadense_Gobar.D06G090700.4 Gobar.D06G090700.4 |
Low | No | 956 | OG0000094 | Get ORF Coding potential... Type: noncoding Potential: -0.921 Folding energies... MFEI: -22.312 AMFE: -0.580 GC content %: 38.494 | |
| Gbarbadense_Gobar.D06G090700.5 Gobar.D06G090700.5 |
Low | No | 932 | OG0000094 | Get ORF Coding potential... Type: noncoding Potential: -0.940 Folding energies... MFEI: -22.167 AMFE: -0.577 GC content %: 38.412 | |
| Gbarbadense_Gobar.D06G090700.6 Gobar.D06G090700.6 |
Low | No | 993 | OG0000094 | Get ORF Coding potential... Type: noncoding Potential: -0.955 Folding energies... MFEI: -23.051 AMFE: -0.596 GC content %: 38.671 | |
| Gbarbadense_Gobar.D06G090700.7 Gobar.D06G090700.7 |
Low | No | 991 | OG0000094 | Get ORF Coding potential... Type: noncoding Potential: -0.795 Folding energies... MFEI: -22.815 AMFE: -0.589 GC content %: 38.749 | |
| Gbarbadense_Gobar.D06G090700.8 Gobar.D06G090700.8 |
Low | No | 1015 | OG0000094 | Get ORF Coding potential... Type: noncoding Potential: -0.800 Folding energies... MFEI: -23.320 AMFE: -0.601 GC content %: 38.818 | |
| Gbarbadense_Gobar.D06G090700.9 Gobar.D06G090700.9 |
Low | No | 1076 | OG0000094 | Get ORF Coding potential... Type: noncoding Potential: -0.812 Folding energies... MFEI: -24.145 AMFE: -0.619 GC content %: 39.033 |
Matches to external databases
| Transcript alias | Database | Version | Hit | Evalue |
|---|---|---|---|---|
| Gbarbadense_Gobar.D06G090700.16 | SwissProt | 2021-04-01 | Q54YG5 | 4.78e-8 |
| Gbarbadense_Gobar.D06G090700.11 | SwissProt | 2021-04-01 | Q94A27 | 4.5e-4 |
| Gbarbadense_Gobar.D06G090700.6 | SwissProt | 2021-04-01 | Q54YG5 | 4.45e-8 |
| Gbarbadense_Gobar.D06G090700.18 | SwissProt | 2021-04-01 | Q94A27 | 1.02e-4 |
| Gbarbadense_Gobar.D06G090700.12 | SwissProt | 2021-04-01 | Q94A27 | 4.44e-4 |
| Gbarbadense_Gobar.D06G090700.7 | SwissProt | 2021-04-01 | Q94A27 | 3.47e-4 |
| Gbarbadense_Gobar.D06G090700.19 | SwissProt | 2021-04-01 | Q54YG5 | 2.33e-9 |
| Gbarbadense_Gobar.D06G090700.13 | SwissProt | 2021-04-01 | Q94A27 | 4.34e-4 |
| Gbarbadense_Gobar.D06G090700.8 | SwissProt | 2021-04-01 | Q8IUR0 | 1.92e-4 |
| Gbarbadense_Gobar.D06G090700.3 | SwissProt | 2021-04-01 | Q54YG5 | 4.04e-9 |
... further results
Gene models