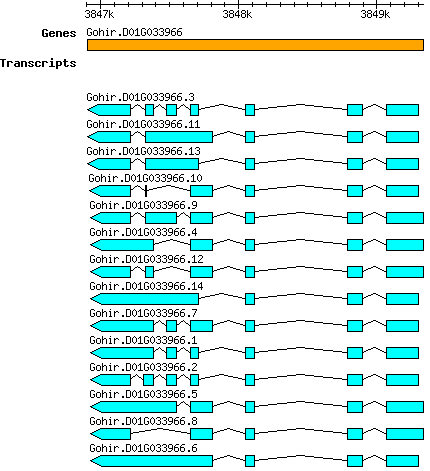
Ghirsutum Gohir.D01G033966
From GreeNC
Gene information
| Gene alias / name | Chrom. | Start | End | Assembly | Species | Details
|
|---|---|---|---|---|---|---|
| Ghirsutum_Gohir.D01G033966 [ Gohir.D01G033966] |
D01 | 3846903 | 3849338 | v2.1 | Gossypium hirsutum (Phytozome 13) | Coding? No |
Transcript features
This table shows how a lncRNA has been classified (high/low confidence) or whether it has been classified as pre-miRNA (go to FAQ for classification criteria). Besides, it displays the lncRNA length and sequence as well as information about the ORF, the coding potential calculated by the Coding Potential Calculator (CPC), and the folding energies calculated by RNAfold.
| Transcript alias / name | Confidence | Pre-miRNA | Length | Orthologous group | Sequence | Details
|
|---|---|---|---|---|---|---|
| Ghirsutum_Gohir.D01G033966.1 Gohir.D01G033966.1 |
Low | No | 1004 | OG0001057 | Get ORF Coding potential... Type: noncoding Potential: -0.752 Folding energies... MFEI: -24.522 AMFE: -0.667 GC content %: 36.753 | |
| Ghirsutum_Gohir.D01G033966.10 Gohir.D01G033966.10 |
Low | No | 879 | OG0001057 | Get ORF Coding potential... Type: noncoding Potential: -0.644 Folding energies... MFEI: -24.596 AMFE: -0.661 GC content %: 37.201 | |
| Ghirsutum_Gohir.D01G033966.11 Gohir.D01G033966.11 |
Low | No | 1213 | OG0001057 | Get ORF Coding potential... Type: noncoding Potential: -0.705 Folding energies... MFEI: -22.795 AMFE: -0.646 GC content %: 35.284 | |
| Ghirsutum_Gohir.D01G033966.12 Gohir.D01G033966.12 |
Low | No | 966 | OG0001057 | Get ORF Coding potential... Type: noncoding Potential: -0.664 Folding energies... MFEI: -23.727 AMFE: -0.631 GC content %: 37.578 | |
| Ghirsutum_Gohir.D01G033966.13 Gohir.D01G033966.13 |
Low | No | 1107 | OG0001057 | Get ORF Coding potential... Type: noncoding Potential: -0.897 Folding energies... MFEI: -22.412 AMFE: -0.641 GC content %: 34.959 | |
| Ghirsutum_Gohir.D01G033966.14 Gohir.D01G033966.14 |
Low | No | 1197 | OG0001057 | Get ORF Coding potential... Type: noncoding Potential: -0.910 Folding energies... MFEI: -23.183 AMFE: -0.654 GC content %: 35.422 | |
| Ghirsutum_Gohir.D01G033966.2 Gohir.D01G033966.2 |
Low | No | 910 | OG0001057 | Get ORF Coding potential... Type: noncoding Potential: -0.729 Folding energies... MFEI: -24.484 AMFE: -0.659 GC content %: 37.143 | |
| Ghirsutum_Gohir.D01G033966.3 Gohir.D01G033966.3 |
Low | No | 915 | OG0001057 | Get ORF Coding potential... Type: noncoding Potential: -0.730 Folding energies... MFEI: -23.792 AMFE: -0.656 GC content %: 36.284 | |
| Ghirsutum_Gohir.D01G033966.4 Gohir.D01G033966.4 |
Low | No | 1074 | OG0001057 | Get ORF Coding potential... Type: noncoding Potential: -0.684 Folding energies... MFEI: -24.348 AMFE: -0.652 GC content %: 37.337 | |
| Ghirsutum_Gohir.D01G033966.5 Gohir.D01G033966.5 |
Low | No | 1243 | OG0001057 | Get ORF Coding potential... Type: noncoding Potential: -0.709 Folding energies... MFEI: -23.105 AMFE: -0.634 GC content %: 36.444 | |
| Ghirsutum_Gohir.D01G033966.6 Gohir.D01G033966.6 |
Low | No | 1303 | OG0001057 | Get ORF Coding potential... Type: noncoding Potential: -0.717 Folding energies... MFEI: -23.254 AMFE: -0.652 GC content %: 35.687 | |
| Ghirsutum_Gohir.D01G033966.7 Gohir.D01G033966.7 |
Low | No | 1110 | OG0001057 | Get ORF Coding potential... Type: noncoding Potential: -0.690 Folding energies... MFEI: -24.279 AMFE: -0.657 GC content %: 36.937 | |
| Ghirsutum_Gohir.D01G033966.8 Gohir.D01G033966.8 |
Low | No | 907 | OG0001057 | Get ORF Coding potential... Type: noncoding Potential: -0.651 Folding energies... MFEI: -23.539 AMFE: -0.626 GC content %: 37.596 | |
| Ghirsutum_Gohir.D01G033966.9 Gohir.D01G033966.9 |
Low | No | 1135 | OG0001057 | Get ORF Coding potential... Type: noncoding Potential: -0.694 Folding energies... MFEI: -22.705 AMFE: -0.621 GC content %: 36.564 |
Matches to external databases
| Transcript alias | Database | Version | Hit | Evalue |
|---|---|---|---|---|
| Ghirsutum_Gohir.D01G033966.2 | SwissProt | 2021-04-01 | B0G107 | 1.43e-8 |
| Ghirsutum_Gohir.D01G033966.10 | SwissProt | 2021-04-01 | Q8S3D1 | 1.34e-4 |
| Ghirsutum_Gohir.D01G033966.8 | SwissProt | 2021-04-01 | Q8S3D1 | 1.45e-4 |
| Ghirsutum_Gohir.D01G033966.3 | SwissProt | 2021-04-01 | Q8S3D1 | 3.16e-5 |
| Ghirsutum_Gohir.D01G033966.11 | SwissProt | 2021-04-01 | Q8S3D1 | 2.52e-4 |
| Ghirsutum_Gohir.D01G033966.9 | SwissProt | 2021-04-01 | B0G107 | 7.22e-7 |
| Ghirsutum_Gohir.D01G033966.4 | SwissProt | 2021-04-01 | Q8S3D1 | 2.02e-4 |
| Ghirsutum_Gohir.D01G033966.12 | SwissProt | 2021-04-01 | Q8S3D1 | 1.65e-4 |
| Ghirsutum_Gohir.D01G033966.5 | SwissProt | 2021-04-01 | Q8S3D1 | 2.61e-4 |
| Ghirsutum_Gohir.D01G033966.13 | SwissProt | 2021-04-01 | Q8S3D1 | 6.06e-5 |
... further results
Gene models