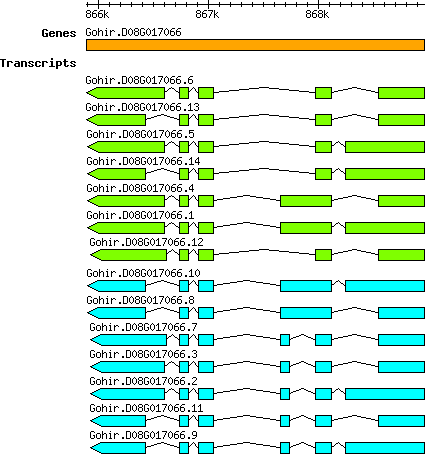
Ghirsutum Gohir.D08G017066
From GreeNC
Gene information
| Gene alias / name | Chrom. | Start | End | Assembly | Species | Details
|
|---|---|---|---|---|---|---|
| Ghirsutum_Gohir.D08G017066 [ Gohir.D08G017066] |
D08 | 865889 | 868966 | v2.1 | Gossypium hirsutum (Phytozome 13) | Coding? No |
Transcript features
This table shows how a lncRNA has been classified (high/low confidence) or whether it has been classified as pre-miRNA (go to FAQ for classification criteria). Besides, it displays the lncRNA length and sequence as well as information about the ORF, the coding potential calculated by the Coding Potential Calculator (CPC), and the folding energies calculated by RNAfold.
| Transcript alias / name | Confidence | Pre-miRNA | Length | Orthologous group | Sequence | Details
|
|---|---|---|---|---|---|---|
| Ghirsutum_Gohir.D08G017066.1 Gohir.D08G017066.1 |
High | No | 2113 | OG0001058 | Get ORF Coding potential... Type: noncoding Potential: -1.098 Folding energies... MFEI: -23.502 AMFE: -0.638 GC content %: 36.820 | |
| Ghirsutum_Gohir.D08G017066.10 Gohir.D08G017066.10 |
Low | No | 1941 | OG0001058 | Get ORF Coding potential... Type: noncoding Potential: -1.083 Folding energies... MFEI: -23.308 AMFE: -0.639 GC content %: 36.476 | |
| Ghirsutum_Gohir.D08G017066.11 Gohir.D08G017066.11 |
Low | No | 1379 | OG0001058 | Get ORF Coding potential... Type: noncoding Potential: -0.245 Folding energies... MFEI: -23.212 AMFE: -0.624 GC content %: 37.201 | |
| Ghirsutum_Gohir.D08G017066.12 Gohir.D08G017066.12 |
High | No | 1489 | OG0001058 | Get ORF Coding potential... Type: noncoding Potential: -1.006 Folding energies... MFEI: -23.647 AMFE: -0.630 GC content %: 37.542 | |
| Ghirsutum_Gohir.D08G017066.13 Gohir.D08G017066.13 |
High | No | 1335 | OG0001058 | Get ORF Coding potential... Type: noncoding Potential: -0.994 Folding energies... MFEI: -23.610 AMFE: -0.637 GC content %: 37.079 | |
| Ghirsutum_Gohir.D08G017066.14 Gohir.D08G017066.14 |
High | No | 1619 | OG0001058 | Get ORF Coding potential... Type: noncoding Potential: -1.042 Folding energies... MFEI: -23.477 AMFE: -0.643 GC content %: 36.504 | |
| Ghirsutum_Gohir.D08G017066.2 Gohir.D08G017066.2 |
Low | No | 1852 | OG0001058 | Get ORF Coding potential... Type: noncoding Potential: -0.387 Folding energies... MFEI: -23.288 AMFE: -0.631 GC content %: 36.933 | |
| Ghirsutum_Gohir.D08G017066.3 Gohir.D08G017066.3 |
Low | No | 1551 | OG0001058 | Get ORF Coding potential... Type: noncoding Potential: -0.264 Folding energies... MFEI: -23.488 AMFE: -0.625 GC content %: 37.589 | |
| Ghirsutum_Gohir.D08G017066.4 Gohir.D08G017066.4 |
High | No | 1812 | OG0001058 | Get ORF Coding potential... Type: noncoding Potential: -0.996 Folding energies... MFEI: -23.703 AMFE: -0.634 GC content %: 37.362 | |
| Ghirsutum_Gohir.D08G017066.5 Gohir.D08G017066.5 |
High | No | 1791 | OG0001058 | Get ORF Coding potential... Type: noncoding Potential: -1.091 Folding energies... MFEI: -23.691 AMFE: -0.642 GC content %: 36.907 | |
| Ghirsutum_Gohir.D08G017066.6 Gohir.D08G017066.6 |
High | No | 1507 | OG0001058 | Get ORF Coding potential... Type: noncoding Potential: -1.007 Folding energies... MFEI: -23.849 AMFE: -0.636 GC content %: 37.492 | |
| Ghirsutum_Gohir.D08G017066.7 Gohir.D08G017066.7 |
Low | No | 1572 | OG0001058 | Get ORF Coding potential... Type: noncoding Potential: -0.265 Folding energies... MFEI: -23.461 AMFE: -0.626 GC content %: 37.468 | |
| Ghirsutum_Gohir.D08G017066.8 Gohir.D08G017066.8 |
Low | No | 1640 | OG0001058 | Get ORF Coding potential... Type: noncoding Potential: -0.987 Folding energies... MFEI: -23.494 AMFE: -0.635 GC content %: 37.012 | |
| Ghirsutum_Gohir.D08G017066.9 Gohir.D08G017066.9 |
Low | No | 1680 | OG0001058 | Get ORF Coding potential... Type: noncoding Potential: -0.378 Folding energies... MFEI: -23.042 AMFE: -0.630 GC content %: 36.548 |
Matches to external databases
| Transcript alias | Database | Version | Hit | Evalue |
|---|---|---|---|---|
| Ghirsutum_Gohir.D08G017066.10 | SwissProt | 2021-04-01 | Q557H3 | 3.09e-4 |
| Ghirsutum_Gohir.D08G017066.9 | SwissProt | 2021-04-01 | A4VCH4 | 7.83e-11 |
| Ghirsutum_Gohir.D08G017066.11 | SwissProt | 2021-04-01 | A4VCH4 | 4.07e-11 |
| Ghirsutum_Gohir.D08G017066.2 | SwissProt | 2021-04-01 | A4VCH4 | 9.07e-11 |
| Ghirsutum_Gohir.D08G017066.3 | SwissProt | 2021-04-01 | A4VCH4 | 5.49e-11 |
| Ghirsutum_Gohir.D08G017066.7 | SwissProt | 2021-04-01 | A4VCH4 | 4.88e-11 |
| Ghirsutum_Gohir.D08G017066.8 | SwissProt | 2021-04-01 | Q557H3 | 2.42e-4 |
Gene models