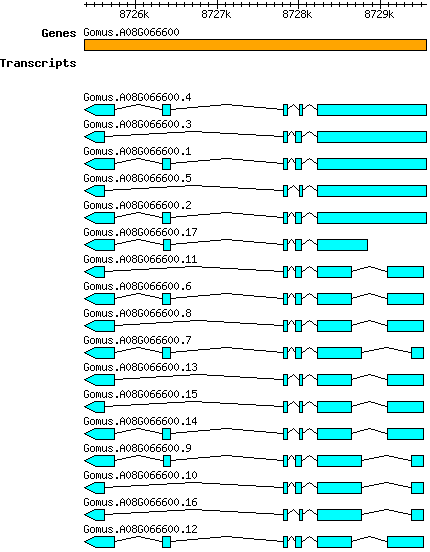
Gmustelinum Gomus.A08G066600
From GreeNC
Gene information
| Gene alias / name | Chrom. | Start | End | Assembly | Species | Details
|
|---|---|---|---|---|---|---|
| Gmustelinum_Gomus.A08G066600 [ Gomus.A08G066600] |
A08 | 8725376 | 8729565 | v1.1 | Gossypium mustelinum (Phytozome 13) | Coding? No |
Transcript features
This table shows how a lncRNA has been classified (high/low confidence) or whether it has been classified as pre-miRNA (go to FAQ for classification criteria). Besides, it displays the lncRNA length and sequence as well as information about the ORF, the coding potential calculated by the Coding Potential Calculator (CPC), and the folding energies calculated by RNAfold.
| Transcript alias / name | Confidence | Pre-miRNA | Length | Orthologous group | Sequence | Details
|
|---|---|---|---|---|---|---|
| Gmustelinum_Gomus.A08G066600.1 Gomus.A08G066600.1 |
Low | No | 1927 | OG0003200 | Get ORF Coding potential... Type: noncoding Potential: -0.267 Folding energies... MFEI: -23.008 AMFE: -0.608 GC content %: 37.831 | |
| Gmustelinum_Gomus.A08G066600.10 Gomus.A08G066600.10 |
Low | No | 1052 | OG0003200 | Get ORF Coding potential... Type: noncoding Potential: -0.215 Folding energies... MFEI: -23.298 AMFE: -0.596 GC content %: 39.068 | |
| Gmustelinum_Gomus.A08G066600.11 Gomus.A08G066600.11 |
Low | No | 1222 | OG0004141 | Get ORF Coding potential... Type: noncoding Potential: -0.228 Folding energies... MFEI: -25.671 AMFE: -0.624 GC content %: 41.162 | |
| Gmustelinum_Gomus.A08G066600.12 Gomus.A08G066600.12 |
Low | No | 1440 | OG0004141 | Get ORF Coding potential... Type: noncoding Potential: -0.247 Folding energies... MFEI: -25.229 AMFE: -0.626 GC content %: 40.278 | |
| Gmustelinum_Gomus.A08G066600.13 Gomus.A08G066600.13 |
Low | No | 1308 | OG0004141 | Get ORF Coding potential... Type: noncoding Potential: -0.439 Folding energies... MFEI: -24.847 AMFE: -0.614 GC content %: 40.443 | |
| Gmustelinum_Gomus.A08G066600.14 Gomus.A08G066600.14 |
Low | No | 1405 | OG0004141 | Get ORF Coding potential... Type: noncoding Potential: -0.447 Folding energies... MFEI: -25.103 AMFE: -0.624 GC content %: 40.214 | |
| Gmustelinum_Gomus.A08G066600.15 Gomus.A08G066600.15 |
Low | No | 1187 | OG0004141 | Get ORF Coding potential... Type: noncoding Potential: -0.428 Folding energies... MFEI: -25.611 AMFE: -0.623 GC content %: 41.112 | |
| Gmustelinum_Gomus.A08G066600.16 Gomus.A08G066600.16 |
Low | No | 1017 | OG0019324 | Get ORF Coding potential... Type: noncoding Potential: -0.409 Folding energies... MFEI: -23.147 AMFE: -0.594 GC content %: 38.938 | |
| Gmustelinum_Gomus.A08G066600.17 Gomus.A08G066600.17 |
Low | No | 1193 | OG0003200 | Get ORF Coding potential... Type: noncoding Potential: -0.216 Folding energies... MFEI: -23.948 AMFE: -0.624 GC content %: 38.391 | |
| Gmustelinum_Gomus.A08G066600.2 Gomus.A08G066600.2 |
Low | No | 1923 | OG0003200 | Get ORF Coding potential... Type: noncoding Potential: -0.266 Folding energies... MFEI: -22.942 AMFE: -0.607 GC content %: 37.806 | |
| Gmustelinum_Gomus.A08G066600.3 Gomus.A08G066600.3 |
Low | No | 1705 | OG0003200 | Get ORF Coding potential... Type: noncoding Potential: -0.276 Folding energies... MFEI: -23.066 AMFE: -0.605 GC content %: 38.123 | |
| Gmustelinum_Gomus.A08G066600.4 Gomus.A08G066600.4 |
Low | No | 1888 | OG0019324 | Get ORF Coding potential... Type: noncoding Potential: -0.474 Folding energies... MFEI: -22.726 AMFE: -0.603 GC content %: 37.712 | |
| Gmustelinum_Gomus.A08G066600.5 Gomus.A08G066600.5 |
Low | No | 1670 | OG0019324 | Get ORF Coding potential... Type: noncoding Potential: -0.467 Folding energies... MFEI: -22.777 AMFE: -0.599 GC content %: 38.024 | |
| Gmustelinum_Gomus.A08G066600.6 Gomus.A08G066600.6 |
Low | No | 1444 | OG0004141 | Get ORF Coding potential... Type: noncoding Potential: -0.247 Folding energies... MFEI: -25.312 AMFE: -0.628 GC content %: 40.305 | |
| Gmustelinum_Gomus.A08G066600.7 Gomus.A08G066600.7 |
Low | No | 1274 | OG0003200 | Get ORF Coding potential... Type: noncoding Potential: -0.226 Folding energies... MFEI: -23.305 AMFE: -0.606 GC content %: 38.462 | |
| Gmustelinum_Gomus.A08G066600.8 Gomus.A08G066600.8 |
Low | No | 1343 | OG0004141 | Get ORF Coding potential... Type: noncoding Potential: -0.228 Folding energies... MFEI: -25.041 AMFE: -0.618 GC content %: 40.506 | |
| Gmustelinum_Gomus.A08G066600.9 Gomus.A08G066600.9 |
Low | No | 1262 | OG0003200 | Get ORF Coding potential... Type: noncoding Potential: -0.225 Folding energies... MFEI: -23.114 AMFE: -0.604 GC content %: 38.273 |
Matches to external databases
| Transcript alias | Database | Version | Hit | Evalue |
|---|---|---|---|---|
| Gmustelinum_Gomus.A08G066600.13 | SwissProt | 2021-04-01 | O00835 | 2.19e-14 |
| Gmustelinum_Gomus.A08G066600.8 | SwissProt | 2021-04-01 | O00835 | 3.06e-17 |
| Gmustelinum_Gomus.A08G066600.3 | SwissProt | 2021-04-01 | O00835 | 6.66e-17 |
| Gmustelinum_Gomus.A08G066600.14 | SwissProt | 2021-04-01 | O00835 | 2.4e-14 |
| Gmustelinum_Gomus.A08G066600.1 | SwissProt | 2021-04-01 | O00835 | 7.21e-17 |
| Gmustelinum_Gomus.A08G066600.9 | SwissProt | 2021-04-01 | O00835 | 4.26e-17 |
| Gmustelinum_Gomus.A08G066600.4 | SwissProt | 2021-04-01 | O00835 | 3.44e-14 |
| Gmustelinum_Gomus.A08G066600.15 | SwissProt | 2021-04-01 | O00835 | 1.93e-14 |
| Gmustelinum_Gomus.A08G066600.10 | SwissProt | 2021-04-01 | O00835 | 3.56e-17 |
| Gmustelinum_Gomus.A08G066600.5 | SwissProt | 2021-04-01 | O00835 | 2.98e-14 |
... further results
Gene models