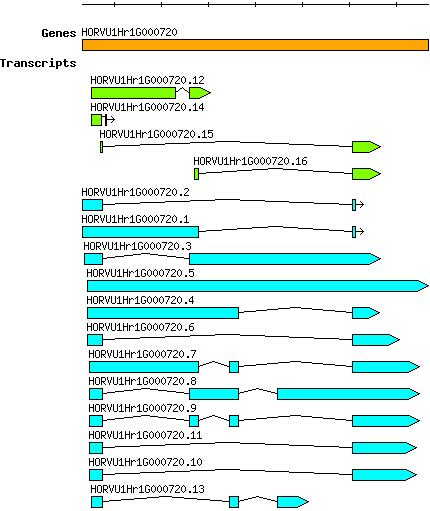
Hvulgare HORVU1Hr1G000720
From GreeNC
Gene information
| Gene alias / name | Chrom. | Start | End | Assembly | Species | Details
|
|---|---|---|---|---|---|---|
| Hvulgare_HORVU1Hr1G000720 [ HORVU1Hr1G000720] |
chr1H | 1922330 | 1929669 | r1 | Hordeum vulgare (Phytozome 13) | Coding? No |
Transcript features
This table shows how a lncRNA has been classified (high/low confidence) or whether it has been classified as pre-miRNA (go to FAQ for classification criteria). Besides, it displays the lncRNA length and sequence as well as information about the ORF, the coding potential calculated by the Coding Potential Calculator (CPC), and the folding energies calculated by RNAfold.
| Transcript alias / name | Confidence | Pre-miRNA | Length | Orthologous group | Sequence | Details
|
|---|---|---|---|---|---|---|
| Hvulgare_HORVU1Hr1G000720.1 HORVU1Hr1G000720.1 |
Low | No | 2527 | OG0012018 | Get ORF Coding potential... Type: noncoding Potential: -1.007 Folding energies... MFEI: -27.277 AMFE: -0.658 GC content %: 41.472 | |
| Hvulgare_HORVU1Hr1G000720.10 HORVU1Hr1G000720.10 |
Low | No | 1640 | OG0012019 | Get ORF Coding potential... Type: noncoding Potential: -0.791 Folding energies... MFEI: -31.543 AMFE: -0.654 GC content %: 48.232 | |
| Hvulgare_HORVU1Hr1G000720.11 HORVU1Hr1G000720.11 |
Low | No | 1639 | OG0012019 | Get ORF Coding potential... Type: noncoding Potential: -0.819 Folding energies... MFEI: -31.544 AMFE: -0.654 GC content %: 48.200 | |
| Hvulgare_HORVU1Hr1G000720.12 HORVU1Hr1G000720.12 |
High | No | 2226 | OG0002320 | Get ORF Coding potential... Type: noncoding Potential: -0.995 Folding energies... MFEI: -27.251 AMFE: -0.659 GC content %: 41.375 | |
| Hvulgare_HORVU1Hr1G000720.13 HORVU1Hr1G000720.13 |
Low | No | 1066 | OG0002320 | Get ORF Coding potential... Type: noncoding Potential: -0.896 Folding energies... MFEI: -25.685 AMFE: -0.563 GC content %: 45.591 | |
| Hvulgare_HORVU1Hr1G000720.14 HORVU1Hr1G000720.14 |
High | No | 217 | OG0002320 | Get ORF Coding potential... Type: noncoding Potential: -0.956 Folding energies... MFEI: -24.378 AMFE: -0.448 GC content %: 54.378 | |
| Hvulgare_HORVU1Hr1G000720.15 HORVU1Hr1G000720.15 |
High | No | 638 | OG0035677 | Get ORF Coding potential... Type: noncoding Potential: -0.547 Folding energies... MFEI: -36.392 AMFE: -0.640 GC content %: 56.897 | |
| Hvulgare_HORVU1Hr1G000720.16 HORVU1Hr1G000720.16 |
High | No | 695 | OG0035678 | Get ORF Coding potential... Type: noncoding Potential: -0.568 Folding energies... MFEI: -34.803 AMFE: -0.622 GC content %: 55.971 | |
| Hvulgare_HORVU1Hr1G000720.2 HORVU1Hr1G000720.2 |
Low | No | 484 | OG0012018 | Get ORF Coding potential... Type: noncoding Potential: -0.712 Folding energies... MFEI: -27.376 AMFE: -0.569 GC content %: 48.140 | |
| Hvulgare_HORVU1Hr1G000720.3 HORVU1Hr1G000720.3 |
Low | No | 4430 | OG0035679 | Get ORF Coding potential... Type: noncoding Potential: -0.837 Folding energies... MFEI: -28.673 AMFE: -0.663 GC content %: 43.228 | |
| Hvulgare_HORVU1Hr1G000720.4 HORVU1Hr1G000720.4 |
Low | No | 3798 | OG0035680 | Get ORF Coding potential... Type: noncoding Potential: -0.832 Folding energies... MFEI: -28.644 AMFE: -0.664 GC content %: 43.154 | |
| Hvulgare_HORVU1Hr1G000720.5 HORVU1Hr1G000720.5 |
Low | No | 7245 | OG0035681 | Get ORF Coding potential... Type: noncoding Potential: -0.848 Folding energies... MFEI: -28.294 AMFE: -0.679 GC content %: 41.684 | |
| Hvulgare_HORVU1Hr1G000720.6 HORVU1Hr1G000720.6 |
Low | No | 1325 | OG0035682 | Get ORF Coding potential... Type: noncoding Potential: -0.775 Folding energies... MFEI: -30.521 AMFE: -0.616 GC content %: 49.509 | |
| Hvulgare_HORVU1Hr1G000720.7 HORVU1Hr1G000720.7 |
Low | No | 3945 | OG0035683 | Get ORF Coding potential... Type: noncoding Potential: -0.833 Folding energies... MFEI: -29.143 AMFE: -0.668 GC content %: 43.625 | |
| Hvulgare_HORVU1Hr1G000720.8 HORVU1Hr1G000720.8 |
Low | No | 4354 | OG0035684 | Get ORF Coding potential... Type: noncoding Potential: -0.836 Folding energies... MFEI: -28.846 AMFE: -0.666 GC content %: 43.294 | |
| Hvulgare_HORVU1Hr1G000720.9 HORVU1Hr1G000720.9 |
Low | No | 2097 | OG0035685 | Get ORF Coding potential... Type: noncoding Potential: -0.806 Folding energies... MFEI: -30.339 AMFE: -0.645 GC content %: 47.020 |
Matches to external databases
| Transcript alias | Database | Version | Hit | Evalue |
|---|---|---|---|---|
| Hvulgare_HORVU1Hr1G000720.7 | RepBase (RepeatMasker) | 2014-01-31 | DNA/CMC-EnSpm; DNA/TcMar-Stowaway; LTR/Copia; SINE/tRNA | |
| Hvulgare_HORVU1Hr1G000720.2 | RepBase (RepeatMasker) | 2014-01-31 | LTR/Copia | |
| Hvulgare_HORVU1Hr1G000720.8 | RepBase (RepeatMasker) | 2014-01-31 | DNA/CMC-EnSpm; DNA/TcMar-Stowaway; LTR/Copia | |
| Hvulgare_HORVU1Hr1G000720.3 | RepBase (RepeatMasker) | 2014-01-31 | DNA/CMC-EnSpm; LTR/Copia | |
| Hvulgare_HORVU1Hr1G000720.1 | RepBase (RepeatMasker) | 2014-01-31 | DNA/CMC-EnSpm; LTR/Copia; SINE/tRNA | |
| Hvulgare_HORVU1Hr1G000720.9 | RepBase (RepeatMasker) | 2014-01-31 | DNA/TcMar-Stowaway; LTR/Copia | |
| Hvulgare_HORVU1Hr1G000720.4 | RepBase (RepeatMasker) | 2014-01-31 | DNA/CMC-EnSpm; LTR/Copia; SINE/tRNA | |
| Hvulgare_HORVU1Hr1G000720.10 | RepBase (RepeatMasker) | 2014-01-31 | DNA/TcMar-Stowaway; LTR/Copia | |
| Hvulgare_HORVU1Hr1G000720.5 | RepBase (RepeatMasker) | 2014-01-31 | DNA/CMC-EnSpm; DNA/TcMar-Stowaway; LTR/Copia; SINE/tRNA | |
| Hvulgare_HORVU1Hr1G000720.11 | RepBase (RepeatMasker) | 2014-01-31 | DNA/TcMar-Stowaway; LTR/Copia |
... further results
Gene models