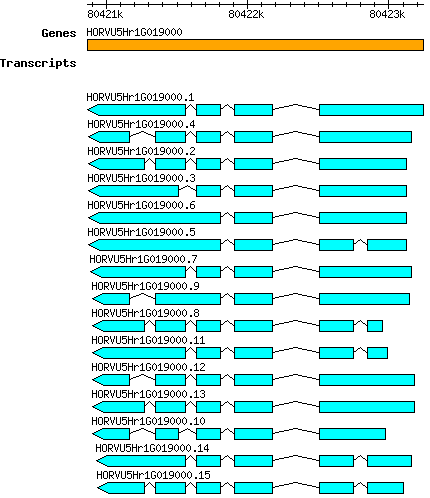
Hvulgare HORVU5Hr1G019000
From GreeNC
Gene information
| Gene alias / name | Chrom. | Start | End | Assembly | Species | Details
|
|---|---|---|---|---|---|---|
| Hvulgare_HORVU5Hr1G019000 [ HORVU5Hr1G019000] |
chr5H | 80420867 | 80423243 | r1 | Hordeum vulgare (Phytozome 13) | Coding? No |
Transcript features
This table shows how a lncRNA has been classified (high/low confidence) or whether it has been classified as pre-miRNA (go to FAQ for classification criteria). Besides, it displays the lncRNA length and sequence as well as information about the ORF, the coding potential calculated by the Coding Potential Calculator (CPC), and the folding energies calculated by RNAfold.
| Transcript alias / name | Confidence | Pre-miRNA | Length | Orthologous group | Sequence | Details
|
|---|---|---|---|---|---|---|
| Hvulgare_HORVU5Hr1G019000.1 HORVU5Hr1G019000.1 |
Low | No | 1866 | OG0000642 | Get ORF Coding potential... Type: noncoding Potential: -0.354 Folding energies... MFEI: -33.215 AMFE: -0.662 GC content %: 50.161 | |
| Hvulgare_HORVU5Hr1G019000.10 HORVU5Hr1G019000.10 |
Low | No | 1334 | OG0044534 | Get ORF Coding potential... Type: noncoding Potential: -1.268 Folding energies... MFEI: -33.973 AMFE: -0.648 GC content %: 52.399 | |
| Hvulgare_HORVU5Hr1G019000.11 HORVU5Hr1G019000.11 |
Low | No | 1480 | OG0044535 | Get ORF Coding potential... Type: noncoding Potential: -0.320 Folding energies... MFEI: -32.378 AMFE: -0.637 GC content %: 50.811 | |
| Hvulgare_HORVU5Hr1G019000.12 HORVU5Hr1G019000.12 |
Low | No | 1582 | OG0000642 | Get ORF Coding potential... Type: noncoding Potential: -1.279 Folding energies... MFEI: -33.799 AMFE: -0.660 GC content %: 51.201 | |
| Hvulgare_HORVU5Hr1G019000.13 HORVU5Hr1G019000.13 |
Low | No | 1696 | OG0000642 | Get ORF Coding potential... Type: noncoding Potential: -1.282 Folding energies... MFEI: -33.974 AMFE: -0.668 GC content %: 50.825 | |
| Hvulgare_HORVU5Hr1G019000.14 HORVU5Hr1G019000.14 |
Low | No | 1625 | OG0012893 | Get ORF Coding potential... Type: noncoding Potential: -0.308 Folding energies... MFEI: -33.249 AMFE: -0.659 GC content %: 50.462 | |
| Hvulgare_HORVU5Hr1G019000.15 HORVU5Hr1G019000.15 |
Low | No | 1487 | OG0044536 | Get ORF Coding potential... Type: noncoding Potential: -1.275 Folding energies... MFEI: -33.753 AMFE: -0.660 GC content %: 51.177 | |
| Hvulgare_HORVU5Hr1G019000.2 HORVU5Hr1G019000.2 |
Low | No | 1664 | OG0000642 | Get ORF Coding potential... Type: noncoding Potential: -1.281 Folding energies... MFEI: -33.612 AMFE: -0.660 GC content %: 50.901 | |
| Hvulgare_HORVU5Hr1G019000.3 HORVU5Hr1G019000.3 |
Low | No | 1693 | OG0000642 | Get ORF Coding potential... Type: noncoding Potential: -0.243 Folding energies... MFEI: -33.024 AMFE: -0.654 GC content %: 50.502 | |
| Hvulgare_HORVU5Hr1G019000.4 HORVU5Hr1G019000.4 |
Low | No | 1599 | OG0000642 | Get ORF Coding potential... Type: noncoding Potential: -1.279 Folding energies... MFEI: -33.652 AMFE: -0.654 GC content %: 51.470 | |
| Hvulgare_HORVU5Hr1G019000.5 HORVU5Hr1G019000.5 |
Low | No | 1713 | OG0012893 | Get ORF Coding potential... Type: noncoding Potential: -0.199 Folding energies... MFEI: -32.119 AMFE: -0.650 GC content %: 49.387 | |
| Hvulgare_HORVU5Hr1G019000.6 HORVU5Hr1G019000.6 |
Low | No | 1813 | OG0000642 | Get ORF Coding potential... Type: noncoding Potential: -0.204 Folding energies... MFEI: -32.427 AMFE: -0.655 GC content %: 49.476 | |
| Hvulgare_HORVU5Hr1G019000.7 HORVU5Hr1G019000.7 |
Low | No | 1769 | OG0000642 | Get ORF Coding potential... Type: noncoding Potential: -0.317 Folding energies... MFEI: -33.103 AMFE: -0.659 GC content %: 50.254 | |
| Hvulgare_HORVU5Hr1G019000.8 HORVU5Hr1G019000.8 |
Low | No | 1365 | OG0044537 | Get ORF Coding potential... Type: noncoding Potential: -1.270 Folding energies... MFEI: -33.377 AMFE: -0.646 GC content %: 51.648 | |
| Hvulgare_HORVU5Hr1G019000.9 HORVU5Hr1G019000.9 |
Low | No | 1625 | OG0000642 | Get ORF Coding potential... Type: noncoding Potential: -1.280 Folding energies... MFEI: -33.169 AMFE: -0.656 GC content %: 50.585 |
Matches to external databases
| Transcript alias | Database | Version | Hit | Evalue |
|---|---|---|---|---|
| Hvulgare_HORVU5Hr1G019000.4 | SwissProt | 2021-04-01 | Q9SZL8 | 1.64e-7 |
| Hvulgare_HORVU5Hr1G019000.6 | RepBase (RepeatMasker) | 2014-01-31 | DNA/hAT-Ac; DNA/MULE-MuDR | |
| Hvulgare_HORVU5Hr1G019000.13 | SwissProt | 2021-04-01 | Q9SZL8 | 7.51e-7 |
| Hvulgare_HORVU5Hr1G019000.15 | RepBase (RepeatMasker) | 2014-01-31 | DNA/hAT-Ac | |
| Hvulgare_HORVU5Hr1G019000.1 | RepBase (RepeatMasker) | 2014-01-31 | DNA/hAT-Ac; DNA/MULE-MuDR | |
| Hvulgare_HORVU5Hr1G019000.5 | SwissProt | 2021-04-01 | Q9SZL8 | 1.72e-18 |
| Hvulgare_HORVU5Hr1G019000.7 | RepBase (RepeatMasker) | 2014-01-31 | DNA/hAT-Ac; DNA/MULE-MuDR | |
| Hvulgare_HORVU5Hr1G019000.14 | SwissProt | 2021-04-01 | Q9SZL8 | 1.76e-16 |
| Hvulgare_HORVU5Hr1G019000.2 | RepBase (RepeatMasker) | 2014-01-31 | DNA/hAT-Ac | |
| Hvulgare_HORVU5Hr1G019000.1 | SwissProt | 2021-04-01 | Q9SZL8 | 7.36e-19 |
... further results
Gene models