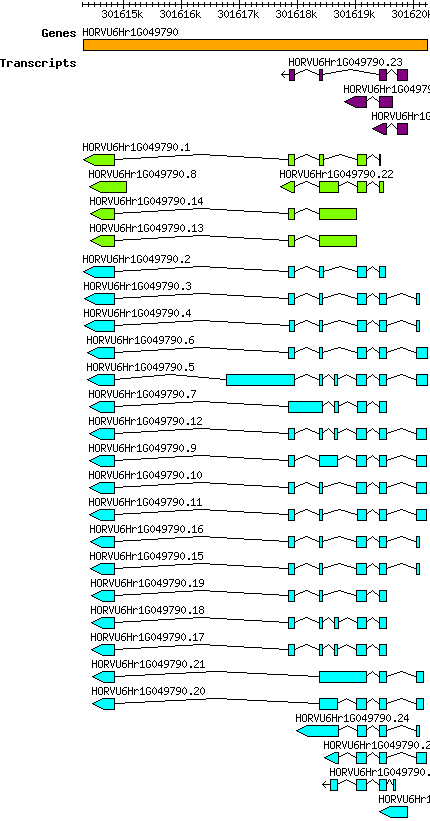
Hvulgare HORVU6Hr1G049790
From GreeNC
Gene information
| Gene alias / name | Chrom. | Start | End | Assembly | Species | Details
|
|---|---|---|---|---|---|---|
| Hvulgare_HORVU6Hr1G049790 [ HORVU6Hr1G049790] |
chr6H | 301614309 | 301620239 | r1 | Hordeum vulgare (Phytozome 13) | Coding? Yes |
Transcript features
This table shows how a lncRNA has been classified (high/low confidence) or whether it has been classified as pre-miRNA (go to FAQ for classification criteria). Besides, it displays the lncRNA length and sequence as well as information about the ORF, the coding potential calculated by the Coding Potential Calculator (CPC), and the folding energies calculated by RNAfold.
| Transcript alias / name | Confidence | Pre-miRNA | Length | Orthologous group | Sequence | Details
|
|---|---|---|---|---|---|---|
| Hvulgare_HORVU6Hr1G049790.1 HORVU6Hr1G049790.1 |
High | No | 882 | OG0047278 | Get ORF Coding potential... Type: noncoding Potential: -1.312 Folding energies... MFEI: -28.277 AMFE: -0.562 GC content %: 50.340 | |
| Hvulgare_HORVU6Hr1G049790.10 HORVU6Hr1G049790.10 |
Low | No | 1034 | OG0005973 | Get ORF Coding potential... Type: noncoding Potential: -0.364 Folding energies... MFEI: -32.650 AMFE: -0.574 GC content %: 56.867 | |
| Hvulgare_HORVU6Hr1G049790.11 HORVU6Hr1G049790.11 |
Low | No | 1034 | OG0005974 | Get ORF Coding potential... Type: noncoding Potential: -0.364 Folding energies... MFEI: -32.708 AMFE: -0.578 GC content %: 56.576 | |
| Hvulgare_HORVU6Hr1G049790.12 HORVU6Hr1G049790.12 |
Low | No | 1110 | OG0005975 | Get ORF Coding potential... Type: noncoding Potential: -0.374 Folding energies... MFEI: -32.198 AMFE: -0.574 GC content %: 56.126 | |
| Hvulgare_HORVU6Hr1G049790.13 HORVU6Hr1G049790.13 |
High | No | 1148 | OG0013152 | Get ORF Coding potential... Type: noncoding Potential: -1.122 Folding energies... MFEI: -26.193 AMFE: -0.578 GC content %: 45.296 | |
| Hvulgare_HORVU6Hr1G049790.14 HORVU6Hr1G049790.14 |
High | No | 1147 | OG0000825 | Get ORF Coding potential... Type: noncoding Potential: -1.122 Folding energies... MFEI: -26.364 AMFE: -0.583 GC content %: 45.248 | |
| Hvulgare_HORVU6Hr1G049790.15 HORVU6Hr1G049790.15 |
Low | No | 892 | OG0000825 | Get ORF Coding potential... Type: noncoding Potential: -0.334 Folding energies... MFEI: -31.211 AMFE: -0.564 GC content %: 55.381 | |
| Hvulgare_HORVU6Hr1G049790.16 HORVU6Hr1G049790.16 |
Low | No | 891 | OG0005973 | Get ORF Coding potential... Type: noncoding Potential: -0.334 Folding energies... MFEI: -30.943 AMFE: -0.559 GC content %: 55.331 | |
| Hvulgare_HORVU6Hr1G049790.17 HORVU6Hr1G049790.17 |
Low | No | 908 | OG0005975 | Get ORF Coding potential... Type: noncoding Potential: -0.346 Folding energies... MFEI: -30.396 AMFE: -0.568 GC content %: 53.524 | |
| Hvulgare_HORVU6Hr1G049790.18 HORVU6Hr1G049790.18 |
Low | No | 909 | OG0005975 | Get ORF Coding potential... Type: noncoding Potential: -0.260 Folding energies... MFEI: -30.143 AMFE: -0.566 GC content %: 53.245 | |
| Hvulgare_HORVU6Hr1G049790.19 HORVU6Hr1G049790.19 |
Low | No | 845 | OG0005973 | Get ORF Coding potential... Type: noncoding Potential: -0.339 Folding energies... MFEI: -30.462 AMFE: -0.562 GC content %: 54.201 | |
| Hvulgare_HORVU6Hr1G049790.2 HORVU6Hr1G049790.2 |
Low | No | 964 | OG0047279 | Get ORF Coding potential... Type: noncoding Potential: -1.130 Folding energies... MFEI: -28.983 AMFE: -0.569 GC content %: 50.934 | |
| Hvulgare_HORVU6Hr1G049790.20 HORVU6Hr1G049790.20 |
Low | No | 1119 | OG0000825 | Get ORF Coding potential... Type: noncoding Potential: -0.367 Folding energies... MFEI: -31.492 AMFE: -0.604 GC content %: 52.100 | |
| Hvulgare_HORVU6Hr1G049790.21 HORVU6Hr1G049790.21 |
Low | No | 1444 | OG0047280 | Get ORF Coding potential... Type: noncoding Potential: -0.395 Folding energies... MFEI: -29.584 AMFE: -0.606 GC content %: 48.823 | |
| Hvulgare_HORVU6Hr1G049790.22 HORVU6Hr1G049790.22 |
High | No | 806 | OG0047281 | Get ORF Coding potential... Type: noncoding Potential: -1.258 Folding energies... MFEI: -24.330 AMFE: -0.524 GC content %: 46.402 | |
| Hvulgare_HORVU6Hr1G049790.24 HORVU6Hr1G049790.24 |
Low | No | 1046 | OG0000825 | Get ORF Coding potential... Type: noncoding Potential: -0.361 Folding energies... MFEI: -24.962 AMFE: -0.571 GC content %: 43.690 | |
| Hvulgare_HORVU6Hr1G049790.25 HORVU6Hr1G049790.25 |
Low | No | 707 | OG0000825 | Get ORF Coding potential... Type: noncoding Potential: -0.289 Folding energies... MFEI: -30.905 AMFE: -0.568 GC content %: 54.455 | |
| Hvulgare_HORVU6Hr1G049790.26 HORVU6Hr1G049790.26 |
Low | No | 465 | OG0000825 | Get ORF Coding potential... Type: noncoding Potential: -0.193 Folding energies... MFEI: -25.699 AMFE: -0.502 GC content %: 51.183 | |
| Hvulgare_HORVU6Hr1G049790.29 HORVU6Hr1G049790.29 |
Low | No | 487 | OG0047282 | Get ORF Coding potential... Type: noncoding Potential: -0.023 Folding energies... MFEI: -31.191 AMFE: -0.548 GC content %: 56.879 | |
| Hvulgare_HORVU6Hr1G049790.3 HORVU6Hr1G049790.3 |
Low | No | 1000 | OG0005974 | Get ORF Coding potential... Type: noncoding Potential: -0.352 Folding energies... MFEI: -30.960 AMFE: -0.583 GC content %: 53.100 | |
| Hvulgare_HORVU6Hr1G049790.4 HORVU6Hr1G049790.4 |
Low | No | 1000 | OG0005974 | Get ORF Coding potential... Type: noncoding Potential: -0.352 Folding energies... MFEI: -30.940 AMFE: -0.583 GC content %: 53.100 | |
| Hvulgare_HORVU6Hr1G049790.5 HORVU6Hr1G049790.5 |
Low | No | 2239 | OG0047283 | Get ORF Coding potential... Type: noncoding Potential: -1.086 Folding energies... MFEI: -27.334 AMFE: -0.575 GC content %: 47.521 | |
| Hvulgare_HORVU6Hr1G049790.6 HORVU6Hr1G049790.6 |
Low | No | 1102 | OG0000825 | Get ORF Coding potential... Type: noncoding Potential: -0.370 Folding energies... MFEI: -32.632 AMFE: -0.587 GC content %: 55.626 | |
| Hvulgare_HORVU6Hr1G049790.7 HORVU6Hr1G049790.7 |
Low | No | 1360 | OG0047284 | Get ORF Coding potential... Type: noncoding Potential: -0.393 Folding energies... MFEI: -26.779 AMFE: -0.565 GC content %: 47.426 | |
| Hvulgare_HORVU6Hr1G049790.8 HORVU6Hr1G049790.8 |
High | No | 633 | OG0013152 | Get ORF Coding potential... Type: noncoding Potential: -1.165 Folding energies... MFEI: -25.940 AMFE: -0.553 GC content %: 46.919 | |
| Hvulgare_HORVU6Hr1G049790.9 HORVU6Hr1G049790.9 |
Low | No | 1305 | OG0000825 | Get ORF Coding potential... Type: noncoding Potential: -1.054 Folding energies... MFEI: -30.751 AMFE: -0.581 GC content %: 52.950 |
Matches to external databases
| Transcript alias | Database | Version | Hit | Evalue |
|---|---|---|---|---|
| Hvulgare_HORVU6Hr1G049790.29 | SwissProt | 2021-04-01 | P15543 | 6.31e-12 |
| Hvulgare_HORVU6Hr1G049790.20 | SwissProt | 2021-04-01 | P15543 | 5.98e-11 |
| Hvulgare_HORVU6Hr1G049790.16 | SwissProt | 2021-04-01 | P15543 | 3.91e-11 |
| Hvulgare_HORVU6Hr1G049790.9 | SwissProt | 2021-04-01 | P15543 | 1.06e-10 |
| Hvulgare_HORVU6Hr1G049790.3 | SwissProt | 2021-04-01 | P15543 | 4.65e-11 |
| Hvulgare_HORVU6Hr1G049790.21 | SwissProt | 2021-04-01 | P15543 | 8.47e-11 |
| Hvulgare_HORVU6Hr1G049790.17 | SwissProt | 2021-04-01 | P15543 | 4.37e-11 |
| Hvulgare_HORVU6Hr1G049790.10 | SwissProt | 2021-04-01 | P15543 | 7.4e-11 |
| Hvulgare_HORVU6Hr1G049790.4 | SwissProt | 2021-04-01 | P15543 | 4.65e-11 |
| Hvulgare_HORVU6Hr1G049790.24 | SwissProt | 2021-04-01 | P15543 | 5.64e-11 |
... further results
Gene models