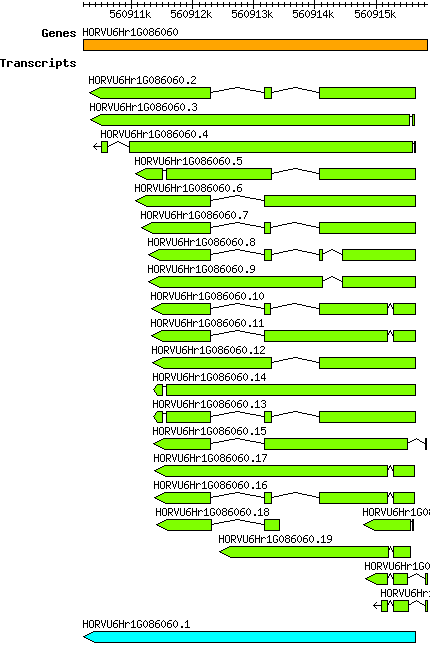
Hvulgare HORVU6Hr1G086060
From GreeNC
Gene information
| Gene alias / name | Chrom. | Start | End | Assembly | Species | Details
|
|---|---|---|---|---|---|---|
| Hvulgare_HORVU6Hr1G086060 [ HORVU6Hr1G086060] |
chr6H | 560910208 | 560915842 | r1 | Hordeum vulgare (Phytozome 13) | Coding? No |
Transcript features
This table shows how a lncRNA has been classified (high/low confidence) or whether it has been classified as pre-miRNA (go to FAQ for classification criteria). Besides, it displays the lncRNA length and sequence as well as information about the ORF, the coding potential calculated by the Coding Potential Calculator (CPC), and the folding energies calculated by RNAfold.
| Transcript alias / name | Confidence | Pre-miRNA | Length | Orthologous group | Sequence | Details
|
|---|---|---|---|---|---|---|
| Hvulgare_HORVU6Hr1G086060.1 HORVU6Hr1G086060.1 |
Low | Yes | 5432 | OG0001436 | Get ORF Coding potential... Type: noncoding Potential: -1.159 Folding energies... MFEI: -29.164 AMFE: -0.690 GC content %: 42.268 | |
| Hvulgare_HORVU6Hr1G086060.10 HORVU6Hr1G086060.10 |
High | No | 2553 | OG0002158 | Get ORF Coding potential... Type: noncoding Potential: -1.234 Folding energies... MFEI: -30.125 AMFE: -0.700 GC content %: 43.008 | |
| Hvulgare_HORVU6Hr1G086060.11 HORVU6Hr1G086060.11 |
High | No | 3352 | OG0019604 | Get ORF Coding potential... Type: noncoding Potential: -1.244 Folding energies... MFEI: -28.303 AMFE: -0.687 GC content %: 41.199 | |
| Hvulgare_HORVU6Hr1G086060.12 HORVU6Hr1G086060.12 |
High | No | 3506 | OG0013224 | Get ORF Coding potential... Type: noncoding Potential: -1.061 Folding energies... MFEI: -29.270 AMFE: -0.694 GC content %: 42.156 | |
| Hvulgare_HORVU6Hr1G086060.13 HORVU6Hr1G086060.13 |
High | No | 2560 | OG0002158 | Get ORF Coding potential... Type: noncoding Potential: -1.234 Folding energies... MFEI: -29.941 AMFE: -0.692 GC content %: 43.242 | |
| Hvulgare_HORVU6Hr1G086060.14 HORVU6Hr1G086060.14 |
High | No | 4227 | OG0001436 | Get ORF Coding potential... Type: noncoding Potential: -1.069 Folding energies... MFEI: -27.845 AMFE: -0.679 GC content %: 40.998 | |
| Hvulgare_HORVU6Hr1G086060.15 HORVU6Hr1G086060.15 |
High | No | 3312 | OG0019604 | Get ORF Coding potential... Type: noncoding Potential: -1.243 Folding energies... MFEI: -27.455 AMFE: -0.685 GC content %: 40.097 | |
| Hvulgare_HORVU6Hr1G086060.16 HORVU6Hr1G086060.16 |
High | No | 2520 | OG0002158 | Get ORF Coding potential... Type: noncoding Potential: -1.233 Folding energies... MFEI: -30.278 AMFE: -0.706 GC content %: 42.897 | |
| Hvulgare_HORVU6Hr1G086060.17 HORVU6Hr1G086060.17 |
High | No | 4187 | OG0001436 | Get ORF Coding potential... Type: noncoding Potential: -1.068 Folding energies... MFEI: -28.013 AMFE: -0.687 GC content %: 40.769 | |
| Hvulgare_HORVU6Hr1G086060.18 HORVU6Hr1G086060.18 |
High | No | 1134 | OG0047997 | Get ORF Coding potential... Type: noncoding Potential: -1.128 Folding energies... MFEI: -27.998 AMFE: -0.635 GC content %: 44.092 | |
| Hvulgare_HORVU6Hr1G086060.19 HORVU6Hr1G086060.19 |
High | No | 3049 | OG0047998 | Get ORF Coding potential... Type: noncoding Potential: -1.185 Folding energies... MFEI: -26.373 AMFE: -0.684 GC content %: 38.537 | |
| Hvulgare_HORVU6Hr1G086060.2 HORVU6Hr1G086060.2 |
High | No | 3665 | OG0047999 | Get ORF Coding potential... Type: noncoding Potential: -1.136 Folding energies... MFEI: -31.181 AMFE: -0.700 GC content %: 44.529 | |
| Hvulgare_HORVU6Hr1G086060.20 HORVU6Hr1G086060.20 |
High | No | 782 | OG0013225 | Get ORF Coding potential... Type: noncoding Potential: -1.256 Folding energies... MFEI: -34.054 AMFE: -0.718 GC content %: 47.442 | |
| Hvulgare_HORVU6Hr1G086060.21 HORVU6Hr1G086060.21 |
High | No | 646 | OG0002158 | Get ORF Coding potential... Type: noncoding Potential: -1.369 Folding energies... MFEI: -34.164 AMFE: -0.736 GC content %: 46.440 | |
| Hvulgare_HORVU6Hr1G086060.22 HORVU6Hr1G086060.22 |
High | No | 379 | OG0048000 | Get ORF Coding potential... Type: noncoding Potential: -1.232 Folding energies... MFEI: -40.079 AMFE: -0.690 GC content %: 58.047 | |
| Hvulgare_HORVU6Hr1G086060.3 HORVU6Hr1G086060.3 |
High | No | 5282 | OG0001436 | Get ORF Coding potential... Type: noncoding Potential: -1.157 Folding energies... MFEI: -28.892 AMFE: -0.686 GC content %: 42.124 | |
| Hvulgare_HORVU6Hr1G086060.4 HORVU6Hr1G086060.4 |
High | No | 4736 | OG0001436 | Get ORF Coding potential... Type: noncoding Potential: -1.027 Folding energies... MFEI: -28.283 AMFE: -0.684 GC content %: 41.322 | |
| Hvulgare_HORVU6Hr1G086060.5 HORVU6Hr1G086060.5 |
High | No | 3730 | OG0013224 | Get ORF Coding potential... Type: noncoding Potential: -1.063 Folding energies... MFEI: -28.992 AMFE: -0.688 GC content %: 42.118 | |
| Hvulgare_HORVU6Hr1G086060.6 HORVU6Hr1G086060.6 |
High | No | 3713 | OG0019604 | Get ORF Coding potential... Type: noncoding Potential: -1.247 Folding energies... MFEI: -27.886 AMFE: -0.679 GC content %: 41.099 | |
| Hvulgare_HORVU6Hr1G086060.7 HORVU6Hr1G086060.7 |
High | No | 2801 | OG0002158 | Get ORF Coding potential... Type: noncoding Potential: -1.238 Folding energies... MFEI: -29.707 AMFE: -0.695 GC content %: 42.770 | |
| Hvulgare_HORVU6Hr1G086060.8 HORVU6Hr1G086060.8 |
High | No | 2365 | OG0013225 | Get ORF Coding potential... Type: noncoding Potential: -1.231 Folding energies... MFEI: -30.605 AMFE: -0.697 GC content %: 43.932 | |
| Hvulgare_HORVU6Hr1G086060.9 HORVU6Hr1G086060.9 |
High | No | 4032 | OG0001436 | Get ORF Coding potential... Type: noncoding Potential: -1.067 Folding energies... MFEI: -28.145 AMFE: -0.682 GC content %: 41.295 |
Matches to external databases
| Transcript alias | Database | Version | Hit | Evalue |
|---|---|---|---|---|
| Hvulgare_HORVU6Hr1G086060.1 | Rfam | 14.6 | MIR1122 | 4.2e-5 |
Gene models