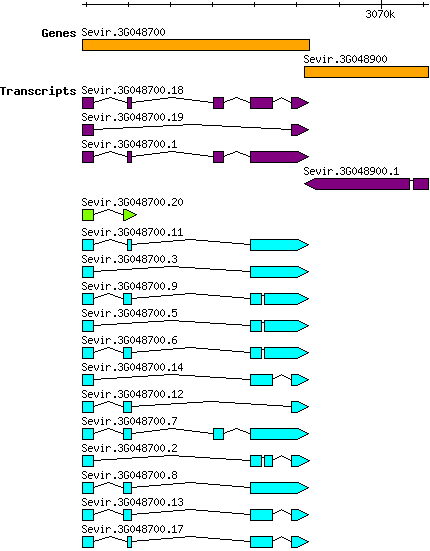
Sviridis Sevir.3G048700
From GreeNC
Gene information
| Gene alias / name | Chrom. | Start | End | Assembly | Species | Details
|
|---|---|---|---|---|---|---|
| Sviridis_Sevir.3G048700 [ Sevir.3G048700] |
Chr_03 | 3062918 | 3068291 | v2.1 | Setaria viridis (Phytozome 13) | Coding? Yes |
Transcript features
This table shows how a lncRNA has been classified (high/low confidence) or whether it has been classified as pre-miRNA (go to FAQ for classification criteria). Besides, it displays the lncRNA length and sequence as well as information about the ORF, the coding potential calculated by the Coding Potential Calculator (CPC), and the folding energies calculated by RNAfold.
| Transcript alias / name | Confidence | Pre-miRNA | Length | Orthologous group | Sequence | Details
|
|---|---|---|---|---|---|---|
| Sviridis_Sevir.3G048700.11 Sevir.3G048700.11 |
Low | No | 1726 | OG0001231 | Get ORF Coding potential... Type: noncoding Potential: -1.061 Folding energies... MFEI: -30.718 AMFE: -0.643 GC content %: 47.798 | |
| Sviridis_Sevir.3G048700.12 Sevir.3G048700.12 |
Low | No | 831 | OG0001231 | Get ORF Coding potential... Type: noncoding Potential: -0.940 Folding energies... MFEI: -30.554 AMFE: -0.607 GC content %: 50.301 | |
| Sviridis_Sevir.3G048700.13 Sevir.3G048700.13 |
Low | No | 1376 | OG0001231 | Get ORF Coding potential... Type: noncoding Potential: -0.989 Folding energies... MFEI: -31.453 AMFE: -0.642 GC content %: 48.983 | |
| Sviridis_Sevir.3G048700.14 Sevir.3G048700.14 |
Low | No | 1194 | OG0001231 | Get ORF Coding potential... Type: noncoding Potential: -0.894 Folding energies... MFEI: -31.432 AMFE: -0.632 GC content %: 49.749 | |
| Sviridis_Sevir.3G048700.17 Sevir.3G048700.17 |
Low | No | 1278 | OG0001231 | Get ORF Coding potential... Type: noncoding Potential: -1.031 Folding energies... MFEI: -31.753 AMFE: -0.633 GC content %: 50.156 | |
| Sviridis_Sevir.3G048700.2 Sevir.3G048700.2 |
Low | No | 1119 | OG0001231 | Get ORF Coding potential... Type: noncoding Potential: -0.836 Folding energies... MFEI: -31.403 AMFE: -0.627 GC content %: 50.045 | |
| Sviridis_Sevir.3G048700.20 Sevir.3G048700.20 |
High | No | 541 | OG0001231 | Get ORF Coding potential... Type: noncoding Potential: -0.887 Folding energies... MFEI: -26.765 AMFE: -0.534 GC content %: 50.092 | |
| Sviridis_Sevir.3G048700.3 Sevir.3G048700.3 |
Low | No | 1625 | OG0001231 | Get ORF Coding potential... Type: noncoding Potential: -0.934 Folding energies... MFEI: -30.351 AMFE: -0.637 GC content %: 47.631 | |
| Sviridis_Sevir.3G048700.5 Sevir.3G048700.5 |
Low | No | 1540 | OG0001231 | Get ORF Coding potential... Type: noncoding Potential: -0.876 Folding energies... MFEI: -30.344 AMFE: -0.634 GC content %: 47.857 | |
| Sviridis_Sevir.3G048700.6 Sevir.3G048700.6 |
Low | No | 1722 | OG0001231 | Get ORF Coding potential... Type: noncoding Potential: -1.005 Folding energies... MFEI: -30.557 AMFE: -0.644 GC content %: 47.445 | |
| Sviridis_Sevir.3G048700.7 Sevir.3G048700.7 |
Low | No | 2037 | OG0001231 | Get ORF Coding potential... Type: noncoding Potential: -1.016 Folding energies... MFEI: -29.593 AMFE: -0.644 GC content %: 45.950 | |
| Sviridis_Sevir.3G048700.8 Sevir.3G048700.8 |
Low | No | 1807 | OG0001231 | Get ORF Coding potential... Type: noncoding Potential: -1.008 Folding energies... MFEI: -30.553 AMFE: -0.646 GC content %: 47.261 | |
| Sviridis_Sevir.3G048700.9 Sevir.3G048700.9 |
Low | No | 1735 | OG0001231 | Get ORF Coding potential... Type: noncoding Potential: -1.006 Folding energies... MFEI: -30.369 AMFE: -0.640 GC content %: 47.435 |
Matches to external databases
| Transcript alias | Database | Version | Hit | Evalue |
|---|---|---|---|---|
| Sviridis_Sevir.3G048700.2 | RepBase (RepeatMasker) | 2014-01-31 | DNA/CMC-EnSpm; DNA/PIF-Harbinger | |
| Sviridis_Sevir.3G048700.11 | RepBase (RepeatMasker) | 2014-01-31 | DNA/CMC-EnSpm; DNA/PIF-Harbinger | |
| Sviridis_Sevir.3G048700.9 | RepBase (RepeatMasker) | 2014-01-31 | DNA/CMC-EnSpm; DNA/PIF-Harbinger | |
| Sviridis_Sevir.3G048700.3 | RepBase (RepeatMasker) | 2014-01-31 | DNA/CMC-EnSpm; DNA/PIF-Harbinger | |
| Sviridis_Sevir.3G048700.12 | RepBase (RepeatMasker) | 2014-01-31 | DNA/CMC-EnSpm; DNA/PIF-Harbinger | |
| Sviridis_Sevir.3G048700.5 | RepBase (RepeatMasker) | 2014-01-31 | DNA/CMC-EnSpm; DNA/PIF-Harbinger | |
| Sviridis_Sevir.3G048700.13 | RepBase (RepeatMasker) | 2014-01-31 | DNA/CMC-EnSpm; DNA/PIF-Harbinger | |
| Sviridis_Sevir.3G048700.6 | RepBase (RepeatMasker) | 2014-01-31 | DNA/CMC-EnSpm; DNA/PIF-Harbinger | |
| Sviridis_Sevir.3G048700.14 | RepBase (RepeatMasker) | 2014-01-31 | DNA/CMC-EnSpm; DNA/PIF-Harbinger | |
| Sviridis_Sevir.3G048700.7 | RepBase (RepeatMasker) | 2014-01-31 | DNA/CMC-EnSpm; DNA/PIF-Harbinger |
... further results
Gene models