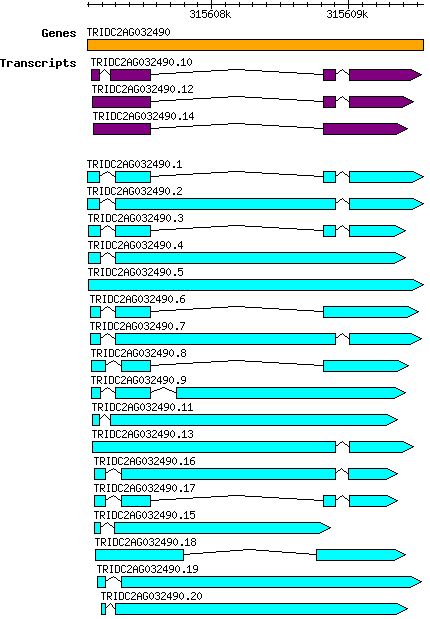
Tdicoccoides TRIDC2AG032490
From GreeNC
Gene information
| Gene alias / name | Chrom. | Start | End | Assembly | Species | Details
|
|---|---|---|---|---|---|---|
| Tdicoccoides_TRIDC2AG032490 [ TRIDC2AG032490] |
2A | 315607092 | 315609547 | v1 | Triticum dicoccoides (Ensembl Plants 51) | Coding? Yes |
Transcript features
This table shows how a lncRNA has been classified (high/low confidence) or whether it has been classified as pre-miRNA (go to FAQ for classification criteria). Besides, it displays the lncRNA length and sequence as well as information about the ORF, the coding potential calculated by the Coding Potential Calculator (CPC), and the folding energies calculated by RNAfold.
| Transcript alias / name | Confidence | Pre-miRNA | Length | Orthologous group | Sequence | Details
|
|---|---|---|---|---|---|---|
| Tdicoccoides_TRIDC2AG032490.1 TRIDC2AG032490.1 |
Low | No | 976 | OG0000739 | Get ORF Coding potential... Type: noncoding Potential: -0.959 Folding energies... MFEI: -31.926 AMFE: -0.644 GC content %: 49.590 | |
| Tdicoccoides_TRIDC2AG032490.11 TRIDC2AG032490.11 |
Low | No | 2150 | OG0000739 | Get ORF Coding potential... Type: noncoding Potential: -1.189 Folding energies... MFEI: -29.265 AMFE: -0.666 GC content %: 43.953 | |
| Tdicoccoides_TRIDC2AG032490.13 TRIDC2AG032490.13 |
Low | No | 2237 | OG0000739 | Get ORF Coding potential... Type: noncoding Potential: -1.067 Folding energies... MFEI: -29.285 AMFE: -0.662 GC content %: 44.256 | |
| Tdicoccoides_TRIDC2AG032490.15 TRIDC2AG032490.15 |
Low | No | 1620 | OG0000739 | Get ORF Coding potential... Type: noncoding Potential: -1.156 Folding energies... MFEI: -28.679 AMFE: -0.646 GC content %: 44.383 | |
| Tdicoccoides_TRIDC2AG032490.16 TRIDC2AG032490.16 |
Low | No | 1994 | OG0000739 | Get ORF Coding potential... Type: noncoding Potential: -1.084 Folding energies... MFEI: -28.831 AMFE: -0.656 GC content %: 43.932 | |
| Tdicoccoides_TRIDC2AG032490.17 TRIDC2AG032490.17 |
Low | No | 725 | OG0000739 | Get ORF Coding potential... Type: noncoding Potential: -0.976 Folding energies... MFEI: -31.972 AMFE: -0.632 GC content %: 50.621 | |
| Tdicoccoides_TRIDC2AG032490.18 TRIDC2AG032490.18 |
Low | No | 1297 | OG0000739 | Get ORF Coding potential... Type: noncoding Potential: -1.009 Folding energies... MFEI: -31.665 AMFE: -0.646 GC content %: 49.036 | |
| Tdicoccoides_TRIDC2AG032490.19 TRIDC2AG032490.19 |
Low | No | 2251 | OG0000739 | Get ORF Coding potential... Type: noncoding Potential: -1.140 Folding energies... MFEI: -28.565 AMFE: -0.666 GC content %: 42.914 | |
| Tdicoccoides_TRIDC2AG032490.2 TRIDC2AG032490.2 |
Low | No | 2239 | OG0000739 | Get ORF Coding potential... Type: noncoding Potential: -1.284 Folding energies... MFEI: -29.201 AMFE: -0.660 GC content %: 44.261 | |
| Tdicoccoides_TRIDC2AG032490.20 TRIDC2AG032490.20 |
Low | No | 2161 | OG0000739 | Get ORF Coding potential... Type: noncoding Potential: -1.171 Folding energies... MFEI: -28.760 AMFE: -0.668 GC content %: 43.082 | |
| Tdicoccoides_TRIDC2AG032490.3 TRIDC2AG032490.3 |
Low | No | 845 | OG0000739 | Get ORF Coding potential... Type: noncoding Potential: -1.051 Folding energies... MFEI: -32.521 AMFE: -0.636 GC content %: 51.124 | |
| Tdicoccoides_TRIDC2AG032490.4 TRIDC2AG032490.4 |
Low | No | 2214 | OG0000739 | Get ORF Coding potential... Type: noncoding Potential: -1.172 Folding energies... MFEI: -29.192 AMFE: -0.662 GC content %: 44.083 | |
| Tdicoccoides_TRIDC2AG032490.5 TRIDC2AG032490.5 |
Low | No | 2449 | OG0000739 | Get ORF Coding potential... Type: noncoding Potential: -1.068 Folding energies... MFEI: -29.367 AMFE: -0.665 GC content %: 44.140 | |
| Tdicoccoides_TRIDC2AG032490.6 TRIDC2AG032490.6 |
Low | No | 1029 | OG0000739 | Get ORF Coding potential... Type: noncoding Potential: -1.123 Folding energies... MFEI: -31.166 AMFE: -0.652 GC content %: 47.813 | |
| Tdicoccoides_TRIDC2AG032490.7 TRIDC2AG032490.7 |
Low | No | 2199 | OG0000739 | Get ORF Coding potential... Type: noncoding Potential: -1.282 Folding energies... MFEI: -28.995 AMFE: -0.659 GC content %: 43.975 | |
| Tdicoccoides_TRIDC2AG032490.8 TRIDC2AG032490.8 |
Low | No | 935 | OG0000739 | Get ORF Coding potential... Type: noncoding Potential: -0.992 Folding energies... MFEI: -30.984 AMFE: -0.644 GC content %: 48.128 | |
| Tdicoccoides_TRIDC2AG032490.9 TRIDC2AG032490.9 |
Low | No | 1998 | OG0000739 | Get ORF Coding potential... Type: noncoding Potential: -1.167 Folding energies... MFEI: -28.649 AMFE: -0.669 GC content %: 42.843 |
Matches to external databases
| Transcript alias | Database | Version | Hit | Evalue |
|---|---|---|---|---|
| Tdicoccoides_TRIDC2AG032490.4 | SwissProt | 2021-04-01 | A2YFN7 | 1.55e-5 |
| Tdicoccoides_TRIDC2AG032490.18 | SwissProt | 2021-04-01 | A2YFN7 | 9.07e-6 |
| Tdicoccoides_TRIDC2AG032490.11 | SwissProt | 2021-04-01 | A2YFN7 | 9.09e-6 |
| Tdicoccoides_TRIDC2AG032490.5 | SwissProt | 2021-04-01 | A2YFN7 | 2.32e-5 |
| Tdicoccoides_TRIDC2AG032490.19 | SwissProt | 2021-04-01 | A2YFN7 | 2.05e-10 |
| Tdicoccoides_TRIDC2AG032490.13 | SwissProt | 2021-04-01 | A2YFN7 | 2.06e-5 |
| Tdicoccoides_TRIDC2AG032490.6 | SwissProt | 2021-04-01 | A2YFN7 | 2.27e-4 |
| Tdicoccoides_TRIDC2AG032490.2 | SwissProt | 2021-04-01 | A2YFN7 | 2.14e-5 |
| Tdicoccoides_TRIDC2AG032490.15 | SwissProt | 2021-04-01 | A2YFN7 | 1.27e-5 |
| Tdicoccoides_TRIDC2AG032490.7 | SwissProt | 2021-04-01 | A2YFN7 | 1.71e-5 |
... further results
Gene models