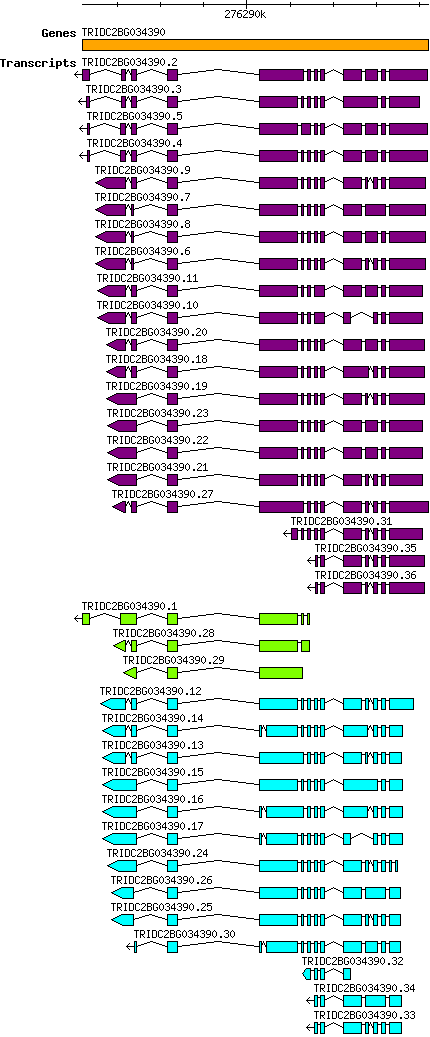
Tdicoccoides TRIDC2BG034390
From GreeNC
Gene information
| Gene alias / name | Chrom. | Start | End | Assembly | Species | Details
|
|---|---|---|---|---|---|---|
| Tdicoccoides_TRIDC2BG034390 [ TRIDC2BG034390] |
2B | 276286205 | 276294198 | v1 | Triticum dicoccoides (Ensembl Plants 51) | Coding? Yes |
Transcript features
This table shows how a lncRNA has been classified (high/low confidence) or whether it has been classified as pre-miRNA (go to FAQ for classification criteria). Besides, it displays the lncRNA length and sequence as well as information about the ORF, the coding potential calculated by the Coding Potential Calculator (CPC), and the folding energies calculated by RNAfold.
| Transcript alias / name | Confidence | Pre-miRNA | Length | Orthologous group | Sequence | Details
|
|---|---|---|---|---|---|---|
| Tdicoccoides_TRIDC2BG034390.1 TRIDC2BG034390.1 |
High | No | 1766 | OG0000742 | Get ORF Coding potential... Type: noncoding Potential: -1.176 Folding energies... MFEI: -29.422 AMFE: -0.657 GC content %: 44.790 | |
| Tdicoccoides_TRIDC2BG034390.12 TRIDC2BG034390.12 |
Low | No | 3333 | OG0000742 | Get ORF Coding potential... Type: noncoding Potential: -1.129 Folding energies... MFEI: -30.393 AMFE: -0.653 GC content %: 46.565 | |
| Tdicoccoides_TRIDC2BG034390.13 TRIDC2BG034390.13 |
Low | No | 3085 | OG0000742 | Get ORF Coding potential... Type: noncoding Potential: -1.126 Folding energies... MFEI: -30.130 AMFE: -0.656 GC content %: 45.932 | |
| Tdicoccoides_TRIDC2BG034390.14 TRIDC2BG034390.14 |
Low | No | 2989 | OG0000742 | Get ORF Coding potential... Type: noncoding Potential: -1.125 Folding energies... MFEI: -30.786 AMFE: -0.663 GC content %: 46.437 | |
| Tdicoccoides_TRIDC2BG034390.15 TRIDC2BG034390.15 |
Low | No | 3360 | OG0000742 | Get ORF Coding potential... Type: noncoding Potential: -1.130 Folding energies... MFEI: -29.655 AMFE: -0.662 GC content %: 44.762 | |
| Tdicoccoides_TRIDC2BG034390.16 TRIDC2BG034390.16 |
Low | No | 3204 | OG0000742 | Get ORF Coding potential... Type: noncoding Potential: -1.128 Folding energies... MFEI: -30.446 AMFE: -0.663 GC content %: 45.943 | |
| Tdicoccoides_TRIDC2BG034390.17 TRIDC2BG034390.17 |
Low | No | 2727 | OG0000742 | Get ORF Coding potential... Type: noncoding Potential: -1.120 Folding energies... MFEI: -30.018 AMFE: -0.660 GC content %: 45.508 | |
| Tdicoccoides_TRIDC2BG034390.24 TRIDC2BG034390.24 |
Low | No | 2837 | OG0000742 | Get ORF Coding potential... Type: noncoding Potential: -1.212 Folding energies... MFEI: -29.778 AMFE: -0.660 GC content %: 45.118 | |
| Tdicoccoides_TRIDC2BG034390.25 TRIDC2BG034390.25 |
Low | No | 2846 | OG0000742 | Get ORF Coding potential... Type: noncoding Potential: -1.122 Folding energies... MFEI: -30.200 AMFE: -0.655 GC content %: 46.135 | |
| Tdicoccoides_TRIDC2BG034390.26 TRIDC2BG034390.26 |
Low | No | 3035 | OG0000742 | Get ORF Coding potential... Type: noncoding Potential: -1.125 Folding energies... MFEI: -29.865 AMFE: -0.657 GC content %: 45.470 | |
| Tdicoccoides_TRIDC2BG034390.28 TRIDC2BG034390.28 |
High | No | 1690 | OG0000742 | Get ORF Coding potential... Type: noncoding Potential: -1.172 Folding energies... MFEI: -28.645 AMFE: -0.633 GC content %: 45.266 | |
| Tdicoccoides_TRIDC2BG034390.29 TRIDC2BG034390.29 |
High | No | 1518 | OG0000742 | Get ORF Coding potential... Type: noncoding Potential: -1.161 Folding energies... MFEI: -28.821 AMFE: -0.643 GC content %: 44.796 | |
| Tdicoccoides_TRIDC2BG034390.30 TRIDC2BG034390.30 |
Low | No | 2383 | OG0000742 | Get ORF Coding potential... Type: noncoding Potential: -1.201 Folding energies... MFEI: -31.326 AMFE: -0.662 GC content %: 47.293 | |
| Tdicoccoides_TRIDC2BG034390.32 TRIDC2BG034390.32 |
Low | No | 481 | OG0000742 | Get ORF Coding potential... Type: noncoding Potential: -0.908 Folding energies... MFEI: -30.104 AMFE: -0.608 GC content %: 49.480 | |
| Tdicoccoides_TRIDC2BG034390.33 TRIDC2BG034390.33 |
Low | No | 1112 | OG0000742 | Get ORF Coding potential... Type: noncoding Potential: -1.048 Folding energies... MFEI: -31.529 AMFE: -0.632 GC content %: 49.910 | |
| Tdicoccoides_TRIDC2BG034390.34 TRIDC2BG034390.34 |
Low | No | 1317 | OG0000742 | Get ORF Coding potential... Type: noncoding Potential: -1.067 Folding energies... MFEI: -30.061 AMFE: -0.631 GC content %: 47.608 |
Matches to external databases
| Transcript alias | Database | Version | Hit | Evalue |
|---|---|---|---|---|
| Tdicoccoides_TRIDC2BG034390.16 | SwissProt | 2021-04-01 | Q6PE01 | 1.28e-7 |
| Tdicoccoides_TRIDC2BG034390.32 | SwissProt | 2021-04-01 | Q9SP22 | 2.88e-5 |
| Tdicoccoides_TRIDC2BG034390.17 | SwissProt | 2021-04-01 | Q10282 | 8.72e-7 |
| Tdicoccoides_TRIDC2BG034390.12 | SwissProt | 2021-04-01 | Q10282 | 1.11e-6 |
| Tdicoccoides_TRIDC2BG034390.33 | SwissProt | 2021-04-01 | Q5RHI5 | 2.17e-7 |
| Tdicoccoides_TRIDC2BG034390.24 | SwissProt | 2021-04-01 | Q10282 | 1.3e-6 |
| Tdicoccoides_TRIDC2BG034390.13 | SwissProt | 2021-04-01 | Q10282 | 1.01e-6 |
| Tdicoccoides_TRIDC2BG034390.34 | SwissProt | 2021-04-01 | A4RJV3 | 1.38e-4 |
| Tdicoccoides_TRIDC2BG034390.25 | SwissProt | 2021-04-01 | Q10282 | 9.18e-7 |
| Tdicoccoides_TRIDC2BG034390.14 | SwissProt | 2021-04-01 | Q6PE01 | 1.17e-7 |
... further results
Gene models