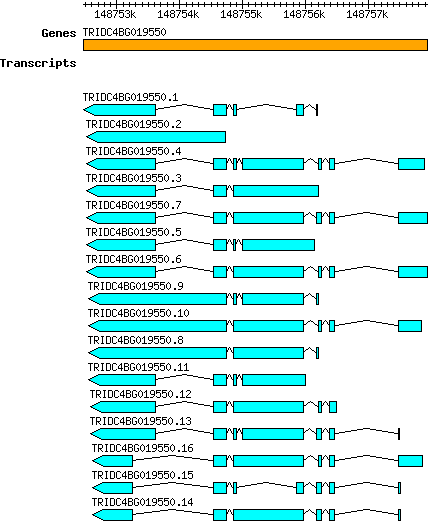
Tdicoccoides TRIDC4BG019550
From GreeNC
Gene information
| Gene alias / name | Chrom. | Start | End | Assembly | Species | Details
|
|---|---|---|---|---|---|---|
| Tdicoccoides_TRIDC4BG019550 [ TRIDC4BG019550] |
4B | 148752470 | 148757936 | v1 | Triticum dicoccoides (Ensembl Plants 51) | Coding? No |
Transcript features
This table shows how a lncRNA has been classified (high/low confidence) or whether it has been classified as pre-miRNA (go to FAQ for classification criteria). Besides, it displays the lncRNA length and sequence as well as information about the ORF, the coding potential calculated by the Coding Potential Calculator (CPC), and the folding energies calculated by RNAfold.
| Transcript alias / name | Confidence | Pre-miRNA | Length | Orthologous group | Sequence | Details
|
|---|---|---|---|---|---|---|
| Tdicoccoides_TRIDC4BG019550.1 TRIDC4BG019550.1 |
Low | No | 1538 | OG0000846 | Get ORF Coding potential... Type: noncoding Potential: -0.064 Folding energies... MFEI: -26.860 AMFE: -0.658 GC content %: 40.832 | |
| Tdicoccoides_TRIDC4BG019550.10 TRIDC4BG019550.10 |
Low | No | 3788 | OG0000846 | Get ORF Coding potential... Type: noncoding Potential: -0.180 Folding energies... MFEI: -28.437 AMFE: -0.687 GC content %: 41.394 | |
| Tdicoccoides_TRIDC4BG019550.11 TRIDC4BG019550.11 |
Low | No | 2306 | OG0000846 | Get ORF Coding potential... Type: noncoding Potential: -0.124 Folding energies... MFEI: -27.095 AMFE: -0.690 GC content %: 39.245 | |
| Tdicoccoides_TRIDC4BG019550.12 TRIDC4BG019550.12 |
Low | No | 2509 | OG0000846 | Get ORF Coding potential... Type: noncoding Potential: -0.131 Folding energies... MFEI: -27.158 AMFE: -0.683 GC content %: 39.777 | |
| Tdicoccoides_TRIDC4BG019550.13 TRIDC4BG019550.13 |
Low | No | 2423 | OG0000846 | Get ORF Coding potential... Type: noncoding Potential: -0.128 Folding energies... MFEI: -27.701 AMFE: -0.681 GC content %: 40.693 | |
| Tdicoccoides_TRIDC4BG019550.14 TRIDC4BG019550.14 |
Low | No | 2151 | OG0000846 | Get ORF Coding potential... Type: noncoding Potential: -0.117 Folding energies... MFEI: -27.931 AMFE: -0.682 GC content %: 40.958 | |
| Tdicoccoides_TRIDC4BG019550.15 TRIDC4BG019550.15 |
Low | No | 1200 | OG0000846 | Get ORF Coding potential... Type: noncoding Potential: -0.007 Folding energies... MFEI: -28.558 AMFE: -0.639 GC content %: 44.667 | |
| Tdicoccoides_TRIDC4BG019550.16 TRIDC4BG019550.16 |
Low | No | 2458 | OG0000846 | Get ORF Coding potential... Type: noncoding Potential: -0.129 Folding energies... MFEI: -30.321 AMFE: -0.684 GC content %: 44.345 | |
| Tdicoccoides_TRIDC4BG019550.2 TRIDC4BG019550.2 |
Low | No | 2225 | OG0000846 | Get ORF Coding potential... Type: noncoding Potential: -0.120 Folding energies... MFEI: -26.157 AMFE: -0.685 GC content %: 38.202 | |
| Tdicoccoides_TRIDC4BG019550.3 TRIDC4BG019550.3 |
Low | No | 2647 | OG0000846 | Get ORF Coding potential... Type: noncoding Potential: -0.156 Folding energies... MFEI: -26.505 AMFE: -0.682 GC content %: 38.836 | |
| Tdicoccoides_TRIDC4BG019550.4 TRIDC4BG019550.4 |
Low | No | 2854 | OG0000846 | Get ORF Coding potential... Type: noncoding Potential: -0.161 Folding energies... MFEI: -30.095 AMFE: -0.685 GC content %: 43.903 | |
| Tdicoccoides_TRIDC4BG019550.5 TRIDC4BG019550.5 |
Low | No | 2487 | OG0000846 | Get ORF Coding potential... Type: noncoding Potential: -0.130 Folding energies... MFEI: -26.803 AMFE: -0.687 GC content %: 39.003 | |
| Tdicoccoides_TRIDC4BG019550.6 TRIDC4BG019550.6 |
Low | No | 2996 | OG0000846 | Get ORF Coding potential... Type: noncoding Potential: -0.165 Folding energies... MFEI: -30.130 AMFE: -0.688 GC content %: 43.825 | |
| Tdicoccoides_TRIDC4BG019550.7 TRIDC4BG019550.7 |
Low | No | 3037 | OG0000846 | Get ORF Coding potential... Type: noncoding Potential: -0.166 Folding energies... MFEI: -30.263 AMFE: -0.687 GC content %: 44.024 | |
| Tdicoccoides_TRIDC4BG019550.8 TRIDC4BG019550.8 |
Low | No | 3337 | OG0000846 | Get ORF Coding potential... Type: noncoding Potential: -0.173 Folding energies... MFEI: -26.440 AMFE: -0.693 GC content %: 38.148 | |
| Tdicoccoides_TRIDC4BG019550.9 TRIDC4BG019550.9 |
Low | No | 3251 | OG0000846 | Get ORF Coding potential... Type: noncoding Potential: -0.171 Folding energies... MFEI: -26.589 AMFE: -0.693 GC content %: 38.357 |
Matches to external databases
| Transcript alias | Database | Version | Hit | Evalue |
|---|---|---|---|---|
| Tdicoccoides_TRIDC4BG019550.7 | SwissProt | 2021-04-01 | Q9FIK0 | 1.83e-15 |
| Tdicoccoides_TRIDC4BG019550.2 | SwissProt | 2021-04-01 | Q9FIK0 | 1.15e-15 |
| Tdicoccoides_TRIDC4BG019550.12 | SwissProt | 2021-04-01 | Q9FIK0 | 1.39e-15 |
| Tdicoccoides_TRIDC4BG019550.8 | SwissProt | 2021-04-01 | Q9FIK0 | 2.09e-15 |
| Tdicoccoides_TRIDC4BG019550.3 | SwissProt | 2021-04-01 | Q9FIK0 | 1.5e-15 |
| Tdicoccoides_TRIDC4BG019550.13 | SwissProt | 2021-04-01 | Q9FIK0 | 1.31e-15 |
| Tdicoccoides_TRIDC4BG019550.9 | SwissProt | 2021-04-01 | Q9FIK0 | 2.02e-15 |
| Tdicoccoides_TRIDC4BG019550.4 | SwissProt | 2021-04-01 | Q9FIK0 | 1.68e-15 |
| Tdicoccoides_TRIDC4BG019550.14 | SwissProt | 2021-04-01 | Q9FIK0 | 1.09e-15 |
| Tdicoccoides_TRIDC4BG019550.1 | SwissProt | 2021-04-01 | Q9FIK0 | 5.59e-16 |
... further results
Gene models