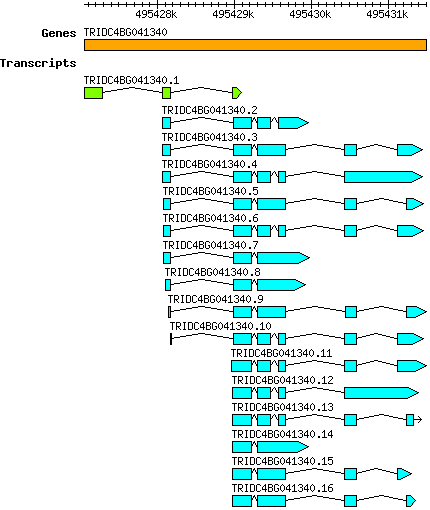
Tdicoccoides TRIDC4BG041340
From GreeNC
Gene information
| Gene alias / name | Chrom. | Start | End | Assembly | Species | Details
|
|---|---|---|---|---|---|---|
| Tdicoccoides_TRIDC4BG041340 [ TRIDC4BG041340] |
4B | 495427050 | 495431500 | v1 | Triticum dicoccoides (Ensembl Plants 51) | Coding? No |
Transcript features
This table shows how a lncRNA has been classified (high/low confidence) or whether it has been classified as pre-miRNA (go to FAQ for classification criteria). Besides, it displays the lncRNA length and sequence as well as information about the ORF, the coding potential calculated by the Coding Potential Calculator (CPC), and the folding energies calculated by RNAfold.
| Transcript alias / name | Confidence | Pre-miRNA | Length | Orthologous group | Sequence | Details
|
|---|---|---|---|---|---|---|
| Tdicoccoides_TRIDC4BG041340.1 TRIDC4BG041340.1 |
High | No | 466 | OG0000200 | Get ORF Coding potential... Type: noncoding Potential: -1.250 Folding energies... MFEI: -33.133 AMFE: -0.506 GC content %: 65.451 | |
| Tdicoccoides_TRIDC4BG041340.10 TRIDC4BG041340.10 |
Low | No | 994 | OG0000200 | Get ORF Coding potential... Type: noncoding Potential: -1.117 Folding energies... MFEI: -27.716 AMFE: -0.636 GC content %: 43.561 | |
| Tdicoccoides_TRIDC4BG041340.11 TRIDC4BG041340.11 |
Low | No | 1044 | OG0000200 | Get ORF Coding potential... Type: noncoding Potential: -1.125 Folding energies... MFEI: -27.385 AMFE: -0.645 GC content %: 42.433 | |
| Tdicoccoides_TRIDC4BG041340.12 TRIDC4BG041340.12 |
Low | No | 1464 | OG0000200 | Get ORF Coding potential... Type: noncoding Potential: -1.176 Folding energies... MFEI: -26.885 AMFE: -0.669 GC content %: 40.164 | |
| Tdicoccoides_TRIDC4BG041340.13 TRIDC4BG041340.13 |
Low | No | 747 | OG0000200 | Get ORF Coding potential... Type: noncoding Potential: -1.060 Folding energies... MFEI: -29.585 AMFE: -0.630 GC content %: 46.988 | |
| Tdicoccoides_TRIDC4BG041340.14 TRIDC4BG041340.14 |
Low | No | 907 | OG0000200 | Get ORF Coding potential... Type: noncoding Potential: -1.100 Folding energies... MFEI: -28.247 AMFE: -0.649 GC content %: 43.550 | |
| Tdicoccoides_TRIDC4BG041340.15 TRIDC4BG041340.15 |
Low | No | 932 | OG0000200 | Get ORF Coding potential... Type: noncoding Potential: -1.105 Folding energies... MFEI: -29.313 AMFE: -0.646 GC content %: 45.386 | |
| Tdicoccoides_TRIDC4BG041340.16 TRIDC4BG041340.16 |
Low | No | 879 | OG0000200 | Get ORF Coding potential... Type: noncoding Potential: -1.094 Folding energies... MFEI: -28.931 AMFE: -0.634 GC content %: 45.620 | |
| Tdicoccoides_TRIDC4BG041340.2 TRIDC4BG041340.2 |
Low | No | 901 | OG0000200 | Get ORF Coding potential... Type: noncoding Potential: -1.098 Folding energies... MFEI: -28.479 AMFE: -0.624 GC content %: 45.616 | |
| Tdicoccoides_TRIDC4BG041340.3 TRIDC4BG041340.3 |
Low | No | 1183 | OG0000200 | Get ORF Coding potential... Type: noncoding Potential: -1.146 Folding energies... MFEI: -28.453 AMFE: -0.639 GC content %: 44.548 | |
| Tdicoccoides_TRIDC4BG041340.4 TRIDC4BG041340.4 |
Low | No | 1620 | OG0000200 | Get ORF Coding potential... Type: noncoding Potential: -1.189 Folding energies... MFEI: -26.833 AMFE: -0.657 GC content %: 40.864 | |
| Tdicoccoides_TRIDC4BG041340.5 TRIDC4BG041340.5 |
Low | No | 1059 | OG0000200 | Get ORF Coding potential... Type: noncoding Potential: -1.128 Folding energies... MFEI: -28.706 AMFE: -0.637 GC content %: 45.042 | |
| Tdicoccoides_TRIDC4BG041340.6 TRIDC4BG041340.6 |
Low | No | 1080 | OG0000200 | Get ORF Coding potential... Type: noncoding Potential: -1.131 Folding energies... MFEI: -28.074 AMFE: -0.632 GC content %: 44.444 | |
| Tdicoccoides_TRIDC4BG041340.7 TRIDC4BG041340.7 |
Low | No | 998 | OG0000200 | Get ORF Coding potential... Type: noncoding Potential: -1.117 Folding energies... MFEI: -28.377 AMFE: -0.631 GC content %: 44.990 | |
| Tdicoccoides_TRIDC4BG041340.8 TRIDC4BG041340.8 |
Low | No | 927 | OG0000200 | Get ORF Coding potential... Type: noncoding Potential: -1.121 Folding energies... MFEI: -28.177 AMFE: -0.625 GC content %: 45.092 | |
| Tdicoccoides_TRIDC4BG041340.9 TRIDC4BG041340.9 |
Low | No | 1030 | OG0000200 | Get ORF Coding potential... Type: noncoding Potential: -1.123 Folding energies... MFEI: -28.068 AMFE: -0.647 GC content %: 43.398 |
Matches to external databases
| Transcript alias | Database | Version | Hit | Evalue |
|---|---|---|---|---|
| Tdicoccoides_TRIDC4BG041340.16 | SwissProt | 2021-04-01 | Q5JK20 | 4.69e-5 |
| Tdicoccoides_TRIDC4BG041340.11 | SwissProt | 2021-04-01 | Q5JK20 | 7.05e-5 |
| Tdicoccoides_TRIDC4BG041340.7 | SwissProt | 2021-04-01 | Q5JK20 | 6.37e-5 |
| Tdicoccoides_TRIDC4BG041340.2 | SwissProt | 2021-04-01 | Q5JK20 | 5.02e-5 |
| Tdicoccoides_TRIDC4BG041340.12 | SwissProt | 2021-04-01 | Q5JK20 | 1.3e-4 |
| Tdicoccoides_TRIDC4BG041340.8 | SwissProt | 2021-04-01 | Q5JK20 | 1.05e-4 |
| Tdicoccoides_TRIDC4BG041340.3 | SwissProt | 2021-04-01 | Q5JK20 | 9.02e-5 |
| Tdicoccoides_TRIDC4BG041340.13 | SwissProt | 2021-04-01 | Q5JK20 | 3.81e-5 |
| Tdicoccoides_TRIDC4BG041340.9 | SwissProt | 2021-04-01 | Q5JK20 | 6.79e-5 |
| Tdicoccoides_TRIDC4BG041340.4 | SwissProt | 2021-04-01 | Q5JK20 | 1.52e-4 |
... further results
Gene models