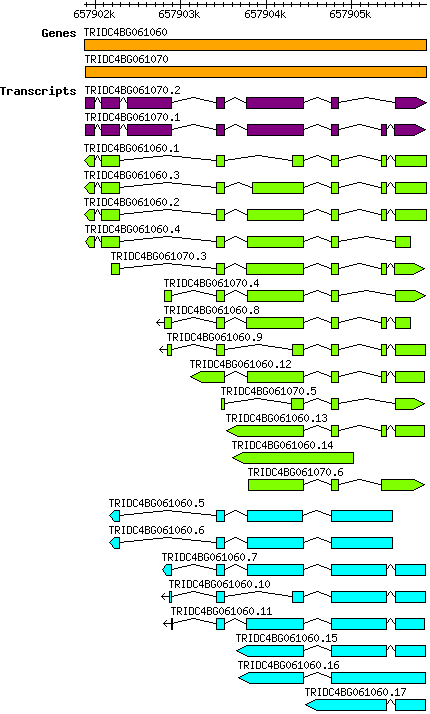
Tdicoccoides TRIDC4BG061060
From GreeNC
Gene information
| Gene alias / name | Chrom. | Start | End | Assembly | Species | Details
|
|---|---|---|---|---|---|---|
| Tdicoccoides_TRIDC4BG061060 [ TRIDC4BG061060] |
4B | 657901876 | 657905872 | v1 | Triticum dicoccoides (Ensembl Plants 51) | Coding? No |
Transcript features
This table shows how a lncRNA has been classified (high/low confidence) or whether it has been classified as pre-miRNA (go to FAQ for classification criteria). Besides, it displays the lncRNA length and sequence as well as information about the ORF, the coding potential calculated by the Coding Potential Calculator (CPC), and the folding energies calculated by RNAfold.
| Transcript alias / name | Confidence | Pre-miRNA | Length | Orthologous group | Sequence | Details
|
|---|---|---|---|---|---|---|
| Tdicoccoides_TRIDC4BG061060.1 TRIDC4BG061060.1 |
High | No | 1033 | OG0000748 | Get ORF Coding potential... Type: noncoding Potential: -1.127 Folding energies... MFEI: -29.487 AMFE: -0.604 GC content %: 48.790 | |
| Tdicoccoides_TRIDC4BG061060.10 TRIDC4BG061060.10 |
Low | No | 1245 | OG0000748 | Get ORF Coding potential... Type: noncoding Potential: -1.090 Folding energies... MFEI: -26.138 AMFE: -0.617 GC content %: 42.329 | |
| Tdicoccoides_TRIDC4BG061060.11 TRIDC4BG061060.11 |
Low | No | 1752 | OG0000748 | Get ORF Coding potential... Type: noncoding Potential: -0.905 Folding energies... MFEI: -24.642 AMFE: -0.594 GC content %: 41.495 | |
| Tdicoccoides_TRIDC4BG061060.12 TRIDC4BG061060.12 |
High | No | 1511 | OG0000748 | Get ORF Coding potential... Type: noncoding Potential: -1.044 Folding energies... MFEI: -24.851 AMFE: -0.602 GC content %: 41.297 | |
| Tdicoccoides_TRIDC4BG061060.13 TRIDC4BG061060.13 |
High | No | 1379 | OG0000748 | Get ORF Coding potential... Type: noncoding Potential: -1.032 Folding energies... MFEI: -24.112 AMFE: -0.586 GC content %: 41.117 | |
| Tdicoccoides_TRIDC4BG061060.14 TRIDC4BG061060.14 |
High | No | 1411 | OG0000748 | Get ORF Coding potential... Type: noncoding Potential: -1.122 Folding energies... MFEI: -23.572 AMFE: -0.592 GC content %: 39.830 | |
| Tdicoccoides_TRIDC4BG061060.15 TRIDC4BG061060.15 |
Low | No | 1781 | OG0000748 | Get ORF Coding potential... Type: noncoding Potential: -1.136 Folding energies... MFEI: -24.003 AMFE: -0.598 GC content %: 40.146 | |
| Tdicoccoides_TRIDC4BG061060.16 TRIDC4BG061060.16 |
Low | No | 1868 | OG0000748 | Get ORF Coding potential... Type: noncoding Potential: -1.141 Folding energies... MFEI: -23.877 AMFE: -0.597 GC content %: 39.989 | |
| Tdicoccoides_TRIDC4BG061060.17 TRIDC4BG061060.17 |
Low | No | 1304 | OG0000748 | Get ORF Coding potential... Type: noncoding Potential: -1.097 Folding energies... MFEI: -23.581 AMFE: -0.602 GC content %: 39.187 | |
| Tdicoccoides_TRIDC4BG061060.2 TRIDC4BG061060.2 |
High | No | 1559 | OG0000748 | Get ORF Coding potential... Type: noncoding Potential: -1.065 Folding energies... MFEI: -28.108 AMFE: -0.609 GC content %: 46.183 | |
| Tdicoccoides_TRIDC4BG061060.3 TRIDC4BG061060.3 |
High | No | 1503 | OG0000748 | Get ORF Coding potential... Type: noncoding Potential: -1.067 Folding energies... MFEI: -27.671 AMFE: -0.600 GC content %: 46.108 | |
| Tdicoccoides_TRIDC4BG061060.4 TRIDC4BG061060.4 |
High | No | 1304 | OG0000748 | Get ORF Coding potential... Type: noncoding Potential: -1.047 Folding energies... MFEI: -29.824 AMFE: -0.629 GC content %: 47.393 | |
| Tdicoccoides_TRIDC4BG061060.5 TRIDC4BG061060.5 |
Low | No | 1544 | OG0000748 | Get ORF Coding potential... Type: noncoding Potential: -1.043 Folding energies... MFEI: -26.192 AMFE: -0.616 GC content %: 42.552 | |
| Tdicoccoides_TRIDC4BG061060.6 TRIDC4BG061060.6 |
Low | No | 1564 | OG0000748 | Get ORF Coding potential... Type: noncoding Potential: -1.121 Folding energies... MFEI: -25.882 AMFE: -0.611 GC content %: 42.391 | |
| Tdicoccoides_TRIDC4BG061060.7 TRIDC4BG061060.7 |
Low | No | 1849 | OG0000748 | Get ORF Coding potential... Type: noncoding Potential: -1.140 Folding energies... MFEI: -26.512 AMFE: -0.612 GC content %: 43.321 | |
| Tdicoccoides_TRIDC4BG061060.8 TRIDC4BG061060.8 |
High | No | 1148 | OG0000748 | Get ORF Coding potential... Type: noncoding Potential: -0.864 Folding energies... MFEI: -28.484 AMFE: -0.626 GC content %: 45.470 | |
| Tdicoccoides_TRIDC4BG061060.9 TRIDC4BG061060.9 |
High | No | 758 | OG0000748 | Get ORF Coding potential... Type: noncoding Potential: -1.093 Folding energies... MFEI: -28.892 AMFE: -0.626 GC content %: 46.174 |
Matches to external databases
| Transcript alias | Database | Version | Hit | Evalue |
|---|---|---|---|---|
| Tdicoccoides_TRIDC4BG061060.5 | SwissProt | 2021-04-01 | Q7XBQ9 | 8.9e-6 |
| Tdicoccoides_TRIDC4BG061060.10 | SwissProt | 2021-04-01 | Q7XBQ9 | 6.07e-6 |
| Tdicoccoides_TRIDC4BG061060.6 | SwissProt | 2021-04-01 | Q7XBQ9 | 9.08e-6 |
| Tdicoccoides_TRIDC4BG061060.11 | SwissProt | 2021-04-01 | Q7XBQ9 | 1.09e-5 |
| Tdicoccoides_TRIDC4BG061060.7 | SwissProt | 2021-04-01 | Q7XBQ9 | 1.18e-5 |
| Tdicoccoides_TRIDC4BG061060.15 | SwissProt | 2021-04-01 | Q7XBQ9 | 1.12e-5 |
| Tdicoccoides_TRIDC4BG061060.16 | SwissProt | 2021-04-01 | Q7XBQ9 | 1.2e-5 |
| Tdicoccoides_TRIDC4BG061060.17 | SwissProt | 2021-04-01 | Q7XBQ9 | 6.61e-6 |
Gene models