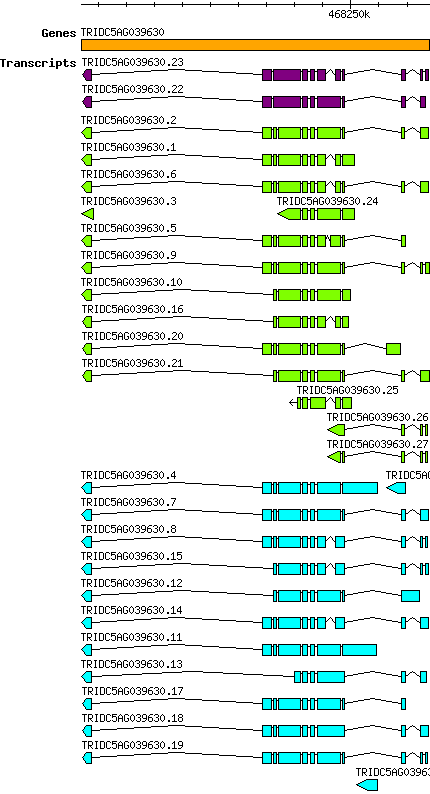
Tdicoccoides TRIDC5AG039630
From GreeNC
Gene information
| Gene alias / name | Chrom. | Start | End | Assembly | Species | Details
|
|---|---|---|---|---|---|---|
| Tdicoccoides_TRIDC5AG039630 [ TRIDC5AG039630] |
5A | 468240418 | 468252784 | v1 | Triticum dicoccoides (Ensembl Plants 51) | Coding? Yes |
Transcript features
This table shows how a lncRNA has been classified (high/low confidence) or whether it has been classified as pre-miRNA (go to FAQ for classification criteria). Besides, it displays the lncRNA length and sequence as well as information about the ORF, the coding potential calculated by the Coding Potential Calculator (CPC), and the folding energies calculated by RNAfold.
| Transcript alias / name | Confidence | Pre-miRNA | Length | Orthologous group | Sequence | Details
|
|---|---|---|---|---|---|---|
| Tdicoccoides_TRIDC5AG039630.1 TRIDC5AG039630.1 |
High | No | 2706 | OG0000264 | Get ORF Coding potential... Type: noncoding Potential: -1.164 Folding energies... MFEI: -30.687 AMFE: -0.637 GC content %: 48.152 | |
| Tdicoccoides_TRIDC5AG039630.10 TRIDC5AG039630.10 |
High | No | 2605 | OG0000264 | Get ORF Coding potential... Type: noncoding Potential: -1.161 Folding energies... MFEI: -27.877 AMFE: -0.633 GC content %: 44.069 | |
| Tdicoccoides_TRIDC5AG039630.11 TRIDC5AG039630.11 |
Low | No | 3848 | OG0000264 | Get ORF Coding potential... Type: noncoding Potential: -1.190 Folding energies... MFEI: -28.495 AMFE: -0.630 GC content %: 45.218 | |
| Tdicoccoides_TRIDC5AG039630.12 TRIDC5AG039630.12 |
Low | No | 3016 | OG0000264 | Get ORF Coding potential... Type: noncoding Potential: -1.173 Folding energies... MFEI: -27.977 AMFE: -0.659 GC content %: 42.440 | |
| Tdicoccoides_TRIDC5AG039630.13 TRIDC5AG039630.13 |
Low | No | 2169 | OG0000264 | Get ORF Coding potential... Type: noncoding Potential: -1.126 Folding energies... MFEI: -29.553 AMFE: -0.647 GC content %: 45.643 | |
| Tdicoccoides_TRIDC5AG039630.14 TRIDC5AG039630.14 |
Low | No | 2849 | OG0000264 | Get ORF Coding potential... Type: noncoding Potential: -1.152 Folding energies... MFEI: -31.162 AMFE: -0.653 GC content %: 47.701 | |
| Tdicoccoides_TRIDC5AG039630.15 TRIDC5AG039630.15 |
Low | No | 2438 | OG0000264 | Get ORF Coding potential... Type: noncoding Potential: -1.138 Folding energies... MFEI: -29.532 AMFE: -0.637 GC content %: 46.349 | |
| Tdicoccoides_TRIDC5AG039630.16 TRIDC5AG039630.16 |
High | No | 2182 | OG0000264 | Get ORF Coding potential... Type: noncoding Potential: -1.144 Folding energies... MFEI: -28.401 AMFE: -0.631 GC content %: 45.005 | |
| Tdicoccoides_TRIDC5AG039630.17 TRIDC5AG039630.17 |
Low | No | 2842 | OG0000264 | Get ORF Coding potential... Type: noncoding Potential: -1.168 Folding energies... MFEI: -29.754 AMFE: -0.652 GC content %: 45.602 | |
| Tdicoccoides_TRIDC5AG039630.18 TRIDC5AG039630.18 |
Low | No | 3213 | OG0000264 | Get ORF Coding potential... Type: noncoding Potential: -1.162 Folding energies... MFEI: -30.383 AMFE: -0.657 GC content %: 46.218 | |
| Tdicoccoides_TRIDC5AG039630.19 TRIDC5AG039630.19 |
Low | No | 2971 | OG0000264 | Get ORF Coding potential... Type: noncoding Potential: -1.156 Folding energies... MFEI: -30.246 AMFE: -0.655 GC content %: 46.146 | |
| Tdicoccoides_TRIDC5AG039630.2 TRIDC5AG039630.2 |
High | No | 3093 | OG0000264 | Get ORF Coding potential... Type: noncoding Potential: -1.175 Folding energies... MFEI: -30.414 AMFE: -0.656 GC content %: 46.363 | |
| Tdicoccoides_TRIDC5AG039630.20 TRIDC5AG039630.20 |
High | No | 3171 | OG0000264 | Get ORF Coding potential... Type: noncoding Potential: -1.177 Folding energies... MFEI: -28.417 AMFE: -0.656 GC content %: 43.330 | |
| Tdicoccoides_TRIDC5AG039630.21 TRIDC5AG039630.21 |
High | No | 2781 | OG0000264 | Get ORF Coding potential... Type: noncoding Potential: -1.167 Folding energies... MFEI: -29.374 AMFE: -0.647 GC content %: 45.379 | |
| Tdicoccoides_TRIDC5AG039630.24 TRIDC5AG039630.24 |
High | No | 2343 | OG0000264 | Get ORF Coding potential... Type: noncoding Potential: -1.151 Folding energies... MFEI: -28.301 AMFE: -0.619 GC content %: 45.753 | |
| Tdicoccoides_TRIDC5AG039630.25 TRIDC5AG039630.25 |
High | No | 1269 | OG0000264 | Get ORF Coding potential... Type: noncoding Potential: -1.133 Folding energies... MFEI: -29.858 AMFE: -0.619 GC content %: 48.227 | |
| Tdicoccoides_TRIDC5AG039630.26 TRIDC5AG039630.26 |
High | No | 875 | OG0000264 | Get ORF Coding potential... Type: noncoding Potential: -1.141 Folding energies... MFEI: -30.217 AMFE: -0.628 GC content %: 48.114 | |
| Tdicoccoides_TRIDC5AG039630.27 TRIDC5AG039630.27 |
High | No | 797 | OG0000264 | Get ORF Coding potential... Type: noncoding Potential: -1.124 Folding energies... MFEI: -30.552 AMFE: -0.618 GC content %: 49.435 | |
| Tdicoccoides_TRIDC5AG039630.28 TRIDC5AG039630.28 |
Low | No | 745 | OG0000264 | Get ORF Coding potential... Type: noncoding Potential: -1.372 Folding energies... MFEI: -16.604 AMFE: -0.460 GC content %: 36.107 | |
| Tdicoccoides_TRIDC5AG039630.29 TRIDC5AG039630.29 |
Low | No | 678 | OG0000264 | Get ORF Coding potential... Type: noncoding Potential: -1.125 Folding energies... MFEI: -27.124 AMFE: -0.648 GC content %: 41.888 | |
| Tdicoccoides_TRIDC5AG039630.3 TRIDC5AG039630.3 |
High | No | 423 | OG0000264 | Get ORF Coding potential... Type: noncoding Potential: -1.171 Folding energies... MFEI: -25.721 AMFE: -0.622 GC content %: 41.371 | |
| Tdicoccoides_TRIDC5AG039630.4 TRIDC5AG039630.4 |
Low | No | 3874 | OG0000264 | Get ORF Coding potential... Type: noncoding Potential: -1.191 Folding energies... MFEI: -28.431 AMFE: -0.630 GC content %: 45.121 | |
| Tdicoccoides_TRIDC5AG039630.5 TRIDC5AG039630.5 |
High | No | 2633 | OG0000264 | Get ORF Coding potential... Type: noncoding Potential: -1.162 Folding energies... MFEI: -30.353 AMFE: -0.650 GC content %: 46.677 | |
| Tdicoccoides_TRIDC5AG039630.6 TRIDC5AG039630.6 |
High | No | 2738 | OG0000264 | Get ORF Coding potential... Type: noncoding Potential: -1.165 Folding energies... MFEI: -31.216 AMFE: -0.651 GC content %: 47.955 | |
| Tdicoccoides_TRIDC5AG039630.7 TRIDC5AG039630.7 |
Low | No | 3140 | OG0000264 | Get ORF Coding potential... Type: noncoding Potential: -1.160 Folding energies... MFEI: -30.627 AMFE: -0.658 GC content %: 46.561 | |
| Tdicoccoides_TRIDC5AG039630.8 TRIDC5AG039630.8 |
Low | No | 2712 | OG0000264 | Get ORF Coding potential... Type: noncoding Potential: -1.148 Folding energies... MFEI: -30.948 AMFE: -0.653 GC content %: 47.419 | |
| Tdicoccoides_TRIDC5AG039630.9 TRIDC5AG039630.9 |
High | No | 3009 | OG0000264 | Get ORF Coding potential... Type: noncoding Potential: -1.173 Folding energies... MFEI: -30.412 AMFE: -0.656 GC content %: 46.394 |
Matches to external databases
| Transcript alias | Database | Version | Hit | Evalue |
|---|---|---|---|---|
| Tdicoccoides_TRIDC5AG039630.29 | RepBase (RepeatMasker) | 2014-01-31 | DNA/hAT-Tip100 | |
| Tdicoccoides_TRIDC5AG039630.15 | RepBase (RepeatMasker) | 2014-01-31 | DNA/hAT-Tip100 | |
| Tdicoccoides_TRIDC5AG039630.4 | RepBase (RepeatMasker) | 2014-01-31 | DNA/CMC-EnSpm | |
| Tdicoccoides_TRIDC5AG039630.17 | RepBase (RepeatMasker) | 2014-01-31 | DNA/hAT-Tip100 | |
| Tdicoccoides_TRIDC5AG039630.11 | RepBase (RepeatMasker) | 2014-01-31 | DNA/CMC-EnSpm | |
| Tdicoccoides_TRIDC5AG039630.7 | RepBase (RepeatMasker) | 2014-01-31 | DNA/hAT-Tip100 | |
| Tdicoccoides_TRIDC5AG039630.18 | RepBase (RepeatMasker) | 2014-01-31 | DNA/hAT-Tip100 | |
| Tdicoccoides_TRIDC5AG039630.12 | RepBase (RepeatMasker) | 2014-01-31 | DNA/hAT-Tip100 | |
| Tdicoccoides_TRIDC5AG039630.8 | RepBase (RepeatMasker) | 2014-01-31 | DNA/hAT-Tip100 | |
| Tdicoccoides_TRIDC5AG039630.19 | RepBase (RepeatMasker) | 2014-01-31 | DNA/hAT-Tip100 |
... further results
Gene models