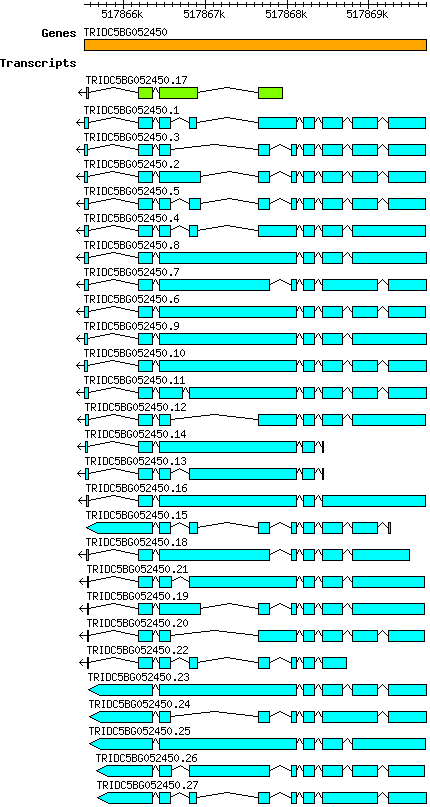
Tdicoccoides TRIDC5BG052450
From GreeNC
Gene information
| Gene alias / name | Chrom. | Start | End | Assembly | Species | Details
|
|---|---|---|---|---|---|---|
| Tdicoccoides_TRIDC5BG052450 [ TRIDC5BG052450] |
5B | 517865528 | 517869701 | v1 | Triticum dicoccoides (Ensembl Plants 51) | Coding? No |
Transcript features
This table shows how a lncRNA has been classified (high/low confidence) or whether it has been classified as pre-miRNA (go to FAQ for classification criteria). Besides, it displays the lncRNA length and sequence as well as information about the ORF, the coding potential calculated by the Coding Potential Calculator (CPC), and the folding energies calculated by RNAfold.
| Transcript alias / name | Confidence | Pre-miRNA | Length | Orthologous group | Sequence | Details
|
|---|---|---|---|---|---|---|
| Tdicoccoides_TRIDC5BG052450.1 TRIDC5BG052450.1 |
Low | No | 2058 | OG0000125 | Get ORF Coding potential... Type: noncoding Potential: -0.562 Folding energies... MFEI: -31.259 AMFE: -0.648 GC content %: 48.251 | |
| Tdicoccoides_TRIDC5BG052450.10 TRIDC5BG052450.10 |
Low | No | 3035 | OG0000125 | Get ORF Coding potential... Type: noncoding Potential: -0.611 Folding energies... MFEI: -28.662 AMFE: -0.658 GC content %: 43.558 | |
| Tdicoccoides_TRIDC5BG052450.11 TRIDC5BG052450.11 |
Low | No | 2966 | OG0000125 | Get ORF Coding potential... Type: noncoding Potential: -0.609 Folding energies... MFEI: -28.759 AMFE: -0.657 GC content %: 43.763 | |
| Tdicoccoides_TRIDC5BG052450.12 TRIDC5BG052450.12 |
Low | No | 2090 | OG0000125 | Get ORF Coding potential... Type: noncoding Potential: -0.564 Folding energies... MFEI: -30.962 AMFE: -0.645 GC content %: 47.990 | |
| Tdicoccoides_TRIDC5BG052450.13 TRIDC5BG052450.13 |
Low | No | 1793 | OG0000125 | Get ORF Coding potential... Type: noncoding Potential: -1.150 Folding energies... MFEI: -25.192 AMFE: -0.634 GC content %: 39.766 | |
| Tdicoccoides_TRIDC5BG052450.14 TRIDC5BG052450.14 |
Low | No | 2011 | OG0000125 | Get ORF Coding potential... Type: noncoding Potential: -1.159 Folding energies... MFEI: -25.037 AMFE: -0.646 GC content %: 38.737 | |
| Tdicoccoides_TRIDC5BG052450.15 TRIDC5BG052450.15 |
Low | No | 1954 | OG0000125 | Get ORF Coding potential... Type: noncoding Potential: -0.554 Folding energies... MFEI: -28.808 AMFE: -0.654 GC content %: 44.063 | |
| Tdicoccoides_TRIDC5BG052450.16 TRIDC5BG052450.16 |
Low | No | 3268 | OG0000125 | Get ORF Coding potential... Type: noncoding Potential: -0.619 Folding energies... MFEI: -28.547 AMFE: -0.665 GC content %: 42.931 | |
| Tdicoccoides_TRIDC5BG052450.17 TRIDC5BG052450.17 |
High | No | 941 | OG0000125 | Get ORF Coding potential... Type: noncoding Potential: -1.087 Folding energies... MFEI: -23.677 AMFE: -0.601 GC content %: 39.426 | |
| Tdicoccoides_TRIDC5BG052450.18 TRIDC5BG052450.18 |
Low | No | 2681 | OG0000125 | Get ORF Coding potential... Type: noncoding Potential: -0.597 Folding energies... MFEI: -28.806 AMFE: -0.680 GC content %: 42.372 | |
| Tdicoccoides_TRIDC5BG052450.19 TRIDC5BG052450.19 |
Low | No | 2163 | OG0000125 | Get ORF Coding potential... Type: noncoding Potential: -0.569 Folding energies... MFEI: -30.865 AMFE: -0.660 GC content %: 46.741 | |
| Tdicoccoides_TRIDC5BG052450.2 TRIDC5BG052450.2 |
Low | No | 2068 | OG0000125 | Get ORF Coding potential... Type: noncoding Potential: -0.562 Folding energies... MFEI: -31.112 AMFE: -0.654 GC content %: 47.582 | |
| Tdicoccoides_TRIDC5BG052450.20 TRIDC5BG052450.20 |
Low | No | 1924 | OG0000125 | Get ORF Coding potential... Type: noncoding Potential: -0.551 Folding energies... MFEI: -30.977 AMFE: -0.637 GC content %: 48.649 | |
| Tdicoccoides_TRIDC5BG052450.21 TRIDC5BG052450.21 |
Low | No | 2905 | OG0000125 | Get ORF Coding potential... Type: noncoding Potential: -0.606 Folding energies... MFEI: -29.143 AMFE: -0.661 GC content %: 44.062 | |
| Tdicoccoides_TRIDC5BG052450.22 TRIDC5BG052450.22 |
Low | No | 1045 | OG0000125 | Get ORF Coding potential... Type: noncoding Potential: -0.429 Folding energies... MFEI: -30.842 AMFE: -0.636 GC content %: 48.517 | |
| Tdicoccoides_TRIDC5BG052450.23 TRIDC5BG052450.23 |
Low | No | 3613 | OG0000125 | Get ORF Coding potential... Type: noncoding Potential: -0.628 Folding energies... MFEI: -28.159 AMFE: -0.665 GC content %: 42.375 | |
| Tdicoccoides_TRIDC5BG052450.24 TRIDC5BG052450.24 |
Low | No | 2375 | OG0000125 | Get ORF Coding potential... Type: noncoding Potential: -0.582 Folding energies... MFEI: -30.535 AMFE: -0.654 GC content %: 46.695 | |
| Tdicoccoides_TRIDC5BG052450.25 TRIDC5BG052450.25 |
Low | No | 3715 | OG0000125 | Get ORF Coding potential... Type: noncoding Potential: -0.631 Folding energies... MFEI: -28.116 AMFE: -0.666 GC content %: 42.207 | |
| Tdicoccoides_TRIDC5BG052450.26 TRIDC5BG052450.26 |
Low | No | 3011 | OG0000125 | Get ORF Coding potential... Type: noncoding Potential: -0.610 Folding energies... MFEI: -29.641 AMFE: -0.670 GC content %: 44.238 | |
| Tdicoccoides_TRIDC5BG052450.27 TRIDC5BG052450.27 |
Low | No | 2369 | OG0000125 | Get ORF Coding potential... Type: noncoding Potential: -0.581 Folding energies... MFEI: -31.368 AMFE: -0.665 GC content %: 47.151 | |
| Tdicoccoides_TRIDC5BG052450.3 TRIDC5BG052450.3 |
Low | No | 1696 | OG0000125 | Get ORF Coding potential... Type: noncoding Potential: -0.530 Folding energies... MFEI: -32.647 AMFE: -0.642 GC content %: 50.825 | |
| Tdicoccoides_TRIDC5BG052450.4 TRIDC5BG052450.4 |
Low | No | 2191 | OG0000125 | Get ORF Coding potential... Type: noncoding Potential: -0.571 Folding energies... MFEI: -31.118 AMFE: -0.654 GC content %: 47.558 | |
| Tdicoccoides_TRIDC5BG052450.5 TRIDC5BG052450.5 |
Low | No | 1848 | OG0000125 | Get ORF Coding potential... Type: noncoding Potential: -0.545 Folding energies... MFEI: -32.570 AMFE: -0.656 GC content %: 49.621 | |
| Tdicoccoides_TRIDC5BG052450.6 TRIDC5BG052450.6 |
Low | No | 3043 | OG0000125 | Get ORF Coding potential... Type: noncoding Potential: -0.611 Folding energies... MFEI: -28.732 AMFE: -0.660 GC content %: 43.510 | |
| Tdicoccoides_TRIDC5BG052450.7 TRIDC5BG052450.7 |
Low | No | 2901 | OG0000125 | Get ORF Coding potential... Type: noncoding Potential: -0.606 Folding energies... MFEI: -29.414 AMFE: -0.670 GC content %: 43.916 | |
| Tdicoccoides_TRIDC5BG052450.8 TRIDC5BG052450.8 |
Low | No | 3174 | OG0000125 | Get ORF Coding potential... Type: noncoding Potential: -0.616 Folding energies... MFEI: -28.781 AMFE: -0.665 GC content %: 43.258 | |
| Tdicoccoides_TRIDC5BG052450.9 TRIDC5BG052450.9 |
Low | No | 3163 | OG0000125 | Get ORF Coding potential... Type: noncoding Potential: -0.615 Folding energies... MFEI: -28.688 AMFE: -0.663 GC content %: 43.282 |
Matches to external databases
| Transcript alias | Database | Version | Hit | Evalue |
|---|---|---|---|---|
| Tdicoccoides_TRIDC5BG052450.20 | SwissProt | 2021-04-01 | Q66GR3 | 5.68e-9 |
| Tdicoccoides_TRIDC5BG052450.15 | SwissProt | 2021-04-01 | Q8H102 | 1.09e-6 |
| Tdicoccoides_TRIDC5BG052450.6 | SwissProt | 2021-04-01 | Q66GR3 | 9.44e-9 |
| Tdicoccoides_TRIDC5BG052450.26 | SwissProt | 2021-04-01 | Q8H102 | 1.92e-6 |
| Tdicoccoides_TRIDC5BG052450.10 | SwissProt | 2021-04-01 | Q66GR3 | 9.4e-9 |
| Tdicoccoides_TRIDC5BG052450.21 | SwissProt | 2021-04-01 | Q66GR3 | 8.96e-9 |
| Tdicoccoides_TRIDC5BG052450.16 | SwissProt | 2021-04-01 | Q66GR3 | 1.02e-8 |
| Tdicoccoides_TRIDC5BG052450.7 | SwissProt | 2021-04-01 | Q8H102 | 1.84e-6 |
| Tdicoccoides_TRIDC5BG052450.27 | SwissProt | 2021-04-01 | Q8H102 | 1.42e-6 |
| Tdicoccoides_TRIDC5BG052450.11 | SwissProt | 2021-04-01 | Q66GR3 | 9.17e-9 |
... further results
Gene models