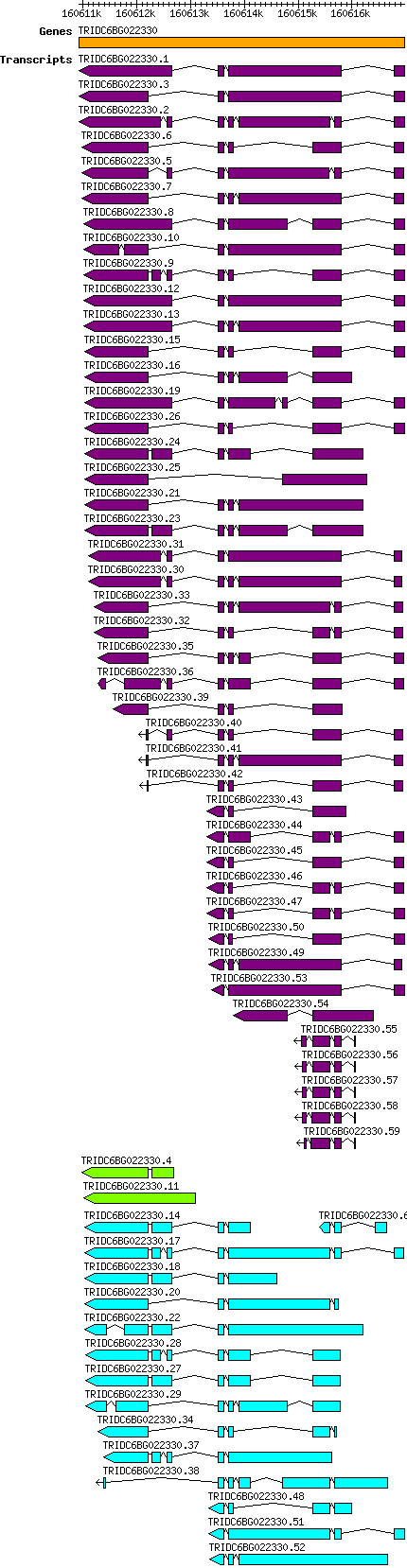
Tdicoccoides TRIDC6BG022330
From GreeNC
Gene information
| Gene alias / name | Chrom. | Start | End | Assembly | Species | Details
|
|---|---|---|---|---|---|---|
| Tdicoccoides_TRIDC6BG022330 [ TRIDC6BG022330] |
6B | 160610927 | 160616978 | v1 | Triticum dicoccoides (Ensembl Plants 51) | Coding? Yes |
Transcript features
This table shows how a lncRNA has been classified (high/low confidence) or whether it has been classified as pre-miRNA (go to FAQ for classification criteria). Besides, it displays the lncRNA length and sequence as well as information about the ORF, the coding potential calculated by the Coding Potential Calculator (CPC), and the folding energies calculated by RNAfold.
| Transcript alias / name | Confidence | Pre-miRNA | Length | Orthologous group | Sequence | Details
|
|---|---|---|---|---|---|---|
| Tdicoccoides_TRIDC6BG022330.11 TRIDC6BG022330.11 |
High | No | 2065 | OG0000853 | Get ORF Coding potential... Type: noncoding Potential: -1.219 Folding energies... MFEI: -24.814 AMFE: -0.614 GC content %: 40.387 | |
| Tdicoccoides_TRIDC6BG022330.14 TRIDC6BG022330.14 |
Low | No | 2065 | OG0000853 | Get ORF Coding potential... Type: noncoding Potential: -0.519 Folding energies... MFEI: -26.504 AMFE: -0.638 GC content %: 41.550 | |
| Tdicoccoides_TRIDC6BG022330.17 TRIDC6BG022330.17 |
Low | No | 3689 | OG0000853 | Get ORF Coding potential... Type: noncoding Potential: -0.074 Folding energies... MFEI: -27.802 AMFE: -0.651 GC content %: 42.722 | |
| Tdicoccoides_TRIDC6BG022330.18 TRIDC6BG022330.18 |
Low | No | 2561 | OG0000853 | Get ORF Coding potential... Type: noncoding Potential: -0.548 Folding energies... MFEI: -25.990 AMFE: -0.629 GC content %: 41.312 | |
| Tdicoccoides_TRIDC6BG022330.20 TRIDC6BG022330.20 |
Low | No | 3219 | OG0000853 | Get ORF Coding potential... Type: noncoding Potential: -0.189 Folding energies... MFEI: -27.465 AMFE: -0.665 GC content %: 41.286 | |
| Tdicoccoides_TRIDC6BG022330.22 TRIDC6BG022330.22 |
Low | No | 3824 | OG0000853 | Get ORF Coding potential... Type: noncoding Potential: -0.029 Folding energies... MFEI: -26.987 AMFE: -0.651 GC content %: 41.449 | |
| Tdicoccoides_TRIDC6BG022330.27 TRIDC6BG022330.27 |
Low | No | 2559 | OG0000853 | Get ORF Coding potential... Type: noncoding Potential: -0.026 Folding energies... MFEI: -28.019 AMFE: -0.657 GC content %: 42.673 | |
| Tdicoccoides_TRIDC6BG022330.28 TRIDC6BG022330.28 |
Low | No | 2425 | OG0000853 | Get ORF Coding potential... Type: noncoding Potential: -0.020 Folding energies... MFEI: -27.868 AMFE: -0.647 GC content %: 43.093 | |
| Tdicoccoides_TRIDC6BG022330.29 TRIDC6BG022330.29 |
Low | No | 2590 | OG0000853 | Get ORF Coding potential... Type: noncoding Potential: -0.054 Folding energies... MFEI: -27.907 AMFE: -0.655 GC content %: 42.587 | |
| Tdicoccoides_TRIDC6BG022330.34 TRIDC6BG022330.34 |
Low | No | 1478 | OG0000853 | Get ORF Coding potential... Type: noncoding Potential: -0.173 Folding energies... MFEI: -27.896 AMFE: -0.627 GC content %: 44.520 | |
| Tdicoccoides_TRIDC6BG022330.37 TRIDC6BG022330.37 |
Low | No | 3079 | OG0000853 | Get ORF Coding potential... Type: noncoding Potential: -0.168 Folding energies... MFEI: -27.223 AMFE: -0.649 GC content %: 41.929 | |
| Tdicoccoides_TRIDC6BG022330.38 TRIDC6BG022330.38 |
Low | No | 2293 | OG0000853 | Get ORF Coding potential... Type: noncoding Potential: -0.156 Folding energies... MFEI: -28.971 AMFE: -0.658 GC content %: 44.047 | |
| Tdicoccoides_TRIDC6BG022330.4 TRIDC6BG022330.4 |
High | No | 1623 | OG0000853 | Get ORF Coding potential... Type: noncoding Potential: -1.191 Folding energies... MFEI: -25.502 AMFE: -0.625 GC content %: 40.789 | |
| Tdicoccoides_TRIDC6BG022330.48 TRIDC6BG022330.48 |
Low | No | 1010 | OG0000853 | Get ORF Coding potential... Type: noncoding Potential: -0.040 Folding energies... MFEI: -27.327 AMFE: -0.643 GC content %: 42.475 | |
| Tdicoccoides_TRIDC6BG022330.51 TRIDC6BG022330.51 |
Low | No | 2462 | OG0000853 | Get ORF Coding potential... Type: noncoding Potential: -0.002 Folding energies... MFEI: -28.168 AMFE: -0.664 GC content %: 42.405 | |
| Tdicoccoides_TRIDC6BG022330.52 TRIDC6BG022330.52 |
Low | No | 3112 | OG0000853 | Get ORF Coding potential... Type: noncoding Potential: -0.012 Folding energies... MFEI: -28.130 AMFE: -0.663 GC content %: 42.416 | |
| Tdicoccoides_TRIDC6BG022330.60 TRIDC6BG022330.60 |
Low | No | 523 | OG0365213 | Get ORF Coding potential... Type: noncoding Potential: -0.045 Folding energies... MFEI: -32.467 AMFE: -0.604 GC content %: 53.728 |
Matches to external databases
| Transcript alias | Database | Version | Hit | Evalue |
|---|---|---|---|---|
| Tdicoccoides_TRIDC6BG022330.37 | SwissProt | 2021-04-01 | Q9LUQ6 | 2.53e-24 |
| Tdicoccoides_TRIDC6BG022330.22 | SwissProt | 2021-04-01 | Q9LUQ6 | 5.11e-38 |
| Tdicoccoides_TRIDC6BG022330.38 | SwissProt | 2021-04-01 | Q9LUQ6 | 3.55e-30 |
| Tdicoccoides_TRIDC6BG022330.27 | SwissProt | 2021-04-01 | Q9LUQ6 | 4.92e-37 |
| Tdicoccoides_TRIDC6BG022330.14 | SwissProt | 2021-04-01 | B4FZ87 | 5.6e-11 |
| Tdicoccoides_TRIDC6BG022330.48 | SwissProt | 2021-04-01 | Q9LUQ6 | 2.62e-30 |
| Tdicoccoides_TRIDC6BG022330.28 | SwissProt | 2021-04-01 | Q9LUQ6 | 4.44e-37 |
| Tdicoccoides_TRIDC6BG022330.17 | SwissProt | 2021-04-01 | Q9LUQ6 | 5.98e-30 |
| Tdicoccoides_TRIDC6BG022330.51 | SwissProt | 2021-04-01 | Q9LUQ6 | 3.89e-30 |
| Tdicoccoides_TRIDC6BG022330.29 | SwissProt | 2021-04-01 | Q9LUQ6 | 1.63e-36 |
... further results
Gene models