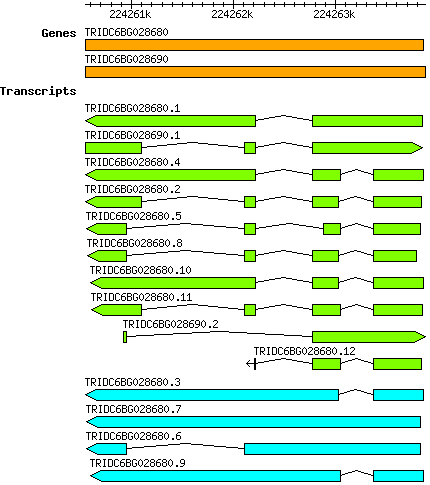
Tdicoccoides TRIDC6BG028680
From GreeNC
Gene information
| Gene alias / name | Chrom. | Start | End | Assembly | Species | Details
|
|---|---|---|---|---|---|---|
| Tdicoccoides_TRIDC6BG028680 [ TRIDC6BG028680] |
6B | 224260550 | 224263859 | v1 | Triticum dicoccoides (Ensembl Plants 51) | Coding? No |
Transcript features
This table shows how a lncRNA has been classified (high/low confidence) or whether it has been classified as pre-miRNA (go to FAQ for classification criteria). Besides, it displays the lncRNA length and sequence as well as information about the ORF, the coding potential calculated by the Coding Potential Calculator (CPC), and the folding energies calculated by RNAfold.
| Transcript alias / name | Confidence | Pre-miRNA | Length | Orthologous group | Sequence | Details
|
|---|---|---|---|---|---|---|
| Tdicoccoides_TRIDC6BG028680.1 TRIDC6BG028680.1 |
High | No | 2746 | OG0001508 | Get ORF Coding potential... Type: noncoding Potential: -1.235 Folding energies... MFEI: -24.487 AMFE: -0.602 GC content %: 40.677 | |
| Tdicoccoides_TRIDC6BG028680.10 TRIDC6BG028680.10 |
High | No | 2363 | OG0001508 | Get ORF Coding potential... Type: noncoding Potential: -1.227 Folding energies... MFEI: -24.757 AMFE: -0.598 GC content %: 41.388 | |
| Tdicoccoides_TRIDC6BG028680.11 TRIDC6BG028680.11 |
High | No | 1359 | OG0001508 | Get ORF Coding potential... Type: noncoding Potential: -1.206 Folding energies... MFEI: -25.372 AMFE: -0.567 GC content %: 44.739 | |
| Tdicoccoides_TRIDC6BG028680.12 TRIDC6BG028680.12 |
High | No | 753 | OG0001508 | Get ORF Coding potential... Type: noncoding Potential: -1.054 Folding energies... MFEI: -15.725 AMFE: -0.395 GC content %: 39.841 | |
| Tdicoccoides_TRIDC6BG028680.2 TRIDC6BG028680.2 |
High | No | 1381 | OG0001508 | Get ORF Coding potential... Type: noncoding Potential: -1.208 Folding energies... MFEI: -25.670 AMFE: -0.574 GC content %: 44.750 | |
| Tdicoccoides_TRIDC6BG028680.3 TRIDC6BG028680.3 |
Low | No | 2965 | OG0001508 | Get ORF Coding potential... Type: noncoding Potential: -1.239 Folding energies... MFEI: -23.730 AMFE: -0.593 GC content %: 40.034 | |
| Tdicoccoides_TRIDC6BG028680.4 TRIDC6BG028680.4 |
High | No | 2424 | OG0001508 | Get ORF Coding potential... Type: noncoding Potential: -1.229 Folding energies... MFEI: -24.748 AMFE: -0.598 GC content %: 41.419 | |
| Tdicoccoides_TRIDC6BG028680.5 TRIDC6BG028680.5 |
High | No | 1123 | OG0001508 | Get ORF Coding potential... Type: noncoding Potential: -1.180 Folding energies... MFEI: -24.853 AMFE: -0.538 GC content %: 46.215 | |
| Tdicoccoides_TRIDC6BG028680.6 TRIDC6BG028680.6 |
Low | No | 2125 | OG0001508 | Get ORF Coding potential... Type: noncoding Potential: -1.252 Folding energies... MFEI: -22.819 AMFE: -0.565 GC content %: 40.376 | |
| Tdicoccoides_TRIDC6BG028680.7 TRIDC6BG028680.7 |
Low | No | 3275 | OG0001508 | Get ORF Coding potential... Type: noncoding Potential: -1.244 Folding energies... MFEI: -23.621 AMFE: -0.595 GC content %: 39.725 | |
| Tdicoccoides_TRIDC6BG028680.8 TRIDC6BG028680.8 |
High | No | 1177 | OG0001508 | Get ORF Coding potential... Type: noncoding Potential: -1.187 Folding energies... MFEI: -25.005 AMFE: -0.556 GC content %: 44.945 | |
| Tdicoccoides_TRIDC6BG028680.9 TRIDC6BG028680.9 |
Low | No | 2934 | OG0001508 | Get ORF Coding potential... Type: noncoding Potential: -1.239 Folding energies... MFEI: -23.712 AMFE: -0.590 GC content %: 40.184 |
Matches to external databases
| Transcript alias | Database | Version | Hit | Evalue |
|---|---|---|---|---|
| Tdicoccoides_TRIDC6BG028680.7 | RepBase (RepeatMasker) | 2014-01-31 | RC/Helitron | |
| Tdicoccoides_TRIDC6BG028680.9 | RepBase (RepeatMasker) | 2014-01-31 | RC/Helitron | |
| Tdicoccoides_TRIDC6BG028680.3 | RepBase (RepeatMasker) | 2014-01-31 | RC/Helitron | |
| Tdicoccoides_TRIDC6BG028680.6 | RepBase (RepeatMasker) | 2014-01-31 | RC/Helitron |
Gene models