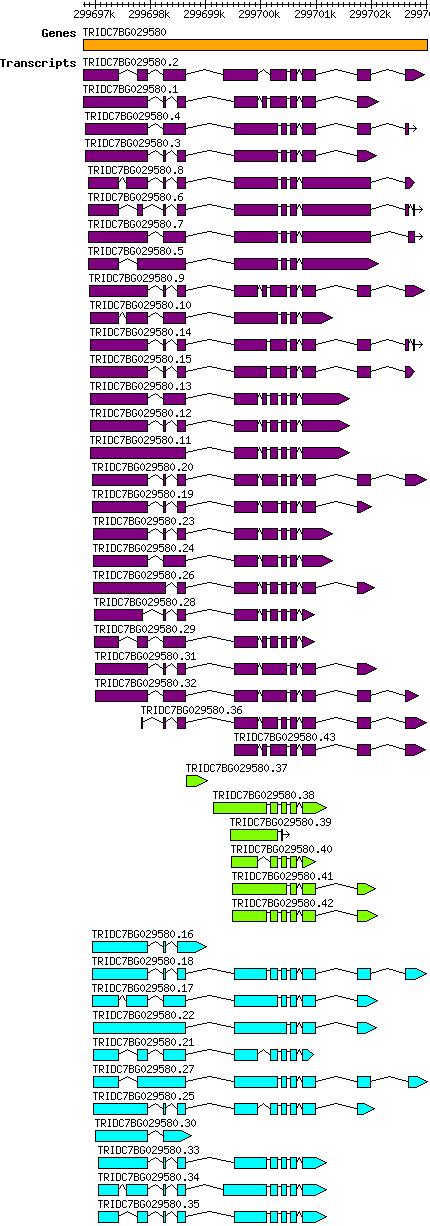
Tdicoccoides TRIDC7BG029580
From GreeNC
Gene information
| Gene alias / name | Chrom. | Start | End | Assembly | Species | Details
|
|---|---|---|---|---|---|---|
| Tdicoccoides_TRIDC7BG029580 [ TRIDC7BG029580] |
7B | 299696783 | 299703022 | v1 | Triticum dicoccoides (Ensembl Plants 51) | Coding? Yes |
Transcript features
This table shows how a lncRNA has been classified (high/low confidence) or whether it has been classified as pre-miRNA (go to FAQ for classification criteria). Besides, it displays the lncRNA length and sequence as well as information about the ORF, the coding potential calculated by the Coding Potential Calculator (CPC), and the folding energies calculated by RNAfold.
| Transcript alias / name | Confidence | Pre-miRNA | Length | Orthologous group | Sequence | Details
|
|---|---|---|---|---|---|---|
| Tdicoccoides_TRIDC7BG029580.16 TRIDC7BG029580.16 |
Low | No | 1570 | OG0000041 | Get ORF Coding potential... Type: noncoding Potential: -0.868 Folding energies... MFEI: -30.159 AMFE: -0.580 GC content %: 51.975 | |
| Tdicoccoides_TRIDC7BG029580.17 TRIDC7BG029580.17 |
Low | No | 2846 | OG0000041 | Get ORF Coding potential... Type: noncoding Potential: -0.938 Folding energies... MFEI: -30.513 AMFE: -0.617 GC content %: 49.438 | |
| Tdicoccoides_TRIDC7BG029580.18 TRIDC7BG029580.18 |
Low | No | 2943 | OG0000041 | Get ORF Coding potential... Type: noncoding Potential: -0.930 Folding energies... MFEI: -30.415 AMFE: -0.622 GC content %: 48.930 | |
| Tdicoccoides_TRIDC7BG029580.21 TRIDC7BG029580.21 |
Low | No | 1996 | OG0000041 | Get ORF Coding potential... Type: noncoding Potential: -0.929 Folding energies... MFEI: -31.273 AMFE: -0.610 GC content %: 51.303 | |
| Tdicoccoides_TRIDC7BG029580.22 TRIDC7BG029580.22 |
Low | No | 3315 | OG0000041 | Get ORF Coding potential... Type: noncoding Potential: -0.973 Folding energies... MFEI: -29.738 AMFE: -0.629 GC content %: 47.270 | |
| Tdicoccoides_TRIDC7BG029580.25 TRIDC7BG029580.25 |
Low | No | 2469 | OG0000041 | Get ORF Coding potential... Type: noncoding Potential: -0.941 Folding energies... MFEI: -31.859 AMFE: -0.622 GC content %: 51.195 | |
| Tdicoccoides_TRIDC7BG029580.27 TRIDC7BG029580.27 |
Low | No | 3139 | OG0000041 | Get ORF Coding potential... Type: noncoding Potential: -0.960 Folding energies... MFEI: -29.573 AMFE: -0.625 GC content %: 47.308 | |
| Tdicoccoides_TRIDC7BG029580.30 TRIDC7BG029580.30 |
Low | No | 1460 | OG0000041 | Get ORF Coding potential... Type: noncoding Potential: -0.981 Folding energies... MFEI: -31.705 AMFE: -0.601 GC content %: 52.740 | |
| Tdicoccoides_TRIDC7BG029580.33 TRIDC7BG029580.33 |
Low | No | 2424 | OG0000041 | Get ORF Coding potential... Type: noncoding Potential: -1.142 Folding energies... MFEI: -30.553 AMFE: -0.634 GC content %: 48.185 | |
| Tdicoccoides_TRIDC7BG029580.34 TRIDC7BG029580.34 |
Low | No | 2488 | OG0000041 | Get ORF Coding potential... Type: noncoding Potential: -1.121 Folding energies... MFEI: -30.631 AMFE: -0.643 GC content %: 47.629 | |
| Tdicoccoides_TRIDC7BG029580.35 TRIDC7BG029580.35 |
Low | No | 2089 | OG0000041 | Get ORF Coding potential... Type: noncoding Potential: -1.124 Folding energies... MFEI: -30.062 AMFE: -0.622 GC content %: 48.301 | |
| Tdicoccoides_TRIDC7BG029580.37 TRIDC7BG029580.37 |
High | No | 385 | OG0000041 | Get ORF Coding potential... Type: noncoding Potential: -1.321 Folding energies... MFEI: -20.364 AMFE: -0.537 GC content %: 37.922 | |
| Tdicoccoides_TRIDC7BG029580.38 TRIDC7BG029580.38 |
High | No | 1715 | OG0000041 | Get ORF Coding potential... Type: noncoding Potential: -1.097 Folding energies... MFEI: -27.283 AMFE: -0.663 GC content %: 41.166 | |
| Tdicoccoides_TRIDC7BG029580.39 TRIDC7BG029580.39 |
High | No | 851 | OG0000041 | Get ORF Coding potential... Type: noncoding Potential: -0.959 Folding energies... MFEI: -24.019 AMFE: -0.614 GC content %: 39.130 | |
| Tdicoccoides_TRIDC7BG029580.40 TRIDC7BG029580.40 |
High | No | 1033 | OG0000041 | Get ORF Coding potential... Type: noncoding Potential: -1.016 Folding energies... MFEI: -28.258 AMFE: -0.639 GC content %: 44.240 | |
| Tdicoccoides_TRIDC7BG029580.41 TRIDC7BG029580.41 |
High | No | 1658 | OG0000041 | Get ORF Coding potential... Type: noncoding Potential: -1.092 Folding energies... MFEI: -27.455 AMFE: -0.638 GC content %: 43.064 | |
| Tdicoccoides_TRIDC7BG029580.42 TRIDC7BG029580.42 |
High | No | 1542 | OG0000041 | Get ORF Coding potential... Type: noncoding Potential: -1.081 Folding energies... MFEI: -28.009 AMFE: -0.639 GC content %: 43.839 |
Matches to external databases
| Transcript alias | Database | Version | Hit | Evalue |
|---|---|---|---|---|
| Tdicoccoides_TRIDC7BG029580.30 | SwissProt | 2021-04-01 | O23614 | 1.61e-6 |
| Tdicoccoides_TRIDC7BG029580.18 | SwissProt | 2021-04-01 | O23614 | 4.41e-6 |
| Tdicoccoides_TRIDC7BG029580.33 | SwissProt | 2021-04-01 | O23614 | 3.43e-6 |
| Tdicoccoides_TRIDC7BG029580.21 | SwissProt | 2021-04-01 | O23614 | 2.71e-7 |
| Tdicoccoides_TRIDC7BG029580.34 | SwissProt | 2021-04-01 | O23614 | 2.33e-6 |
| Tdicoccoides_TRIDC7BG029580.22 | SwissProt | 2021-04-01 | O23614 | 5.12e-6 |
| Tdicoccoides_TRIDC7BG029580.35 | SwissProt | 2021-04-01 | O23614 | 5.3e-7 |
| Tdicoccoides_TRIDC7BG029580.25 | SwissProt | 2021-04-01 | O23614 | 3.52e-6 |
| Tdicoccoides_TRIDC7BG029580.16 | SwissProt | 2021-04-01 | O23614 | 1.82e-6 |
| Tdicoccoides_TRIDC7BG029580.18 | RepBase (RepeatMasker) | 2014-01-31 | LINE/L1 |
... further results
Gene models