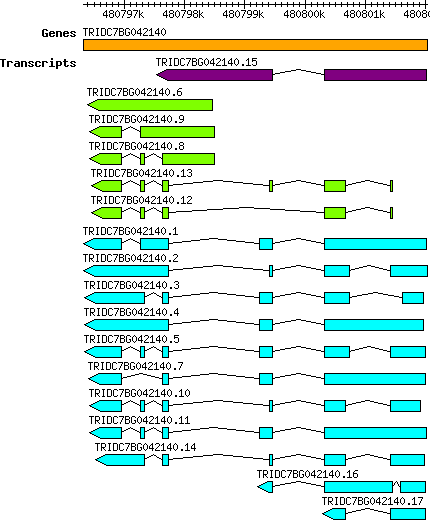
Tdicoccoides TRIDC7BG042140
From GreeNC
Gene information
| Gene alias / name | Chrom. | Start | End | Assembly | Species | Details
|
|---|---|---|---|---|---|---|
| Tdicoccoides_TRIDC7BG042140 [ TRIDC7BG042140] |
7B | 480796326 | 480802026 | v1 | Triticum dicoccoides (Ensembl Plants 51) | Coding? Yes |
Transcript features
This table shows how a lncRNA has been classified (high/low confidence) or whether it has been classified as pre-miRNA (go to FAQ for classification criteria). Besides, it displays the lncRNA length and sequence as well as information about the ORF, the coding potential calculated by the Coding Potential Calculator (CPC), and the folding energies calculated by RNAfold.
| Transcript alias / name | Confidence | Pre-miRNA | Length | Orthologous group | Sequence | Details
|
|---|---|---|---|---|---|---|
| Tdicoccoides_TRIDC7BG042140.1 TRIDC7BG042140.1 |
Low | No | 3005 | OG0000972 | Get ORF Coding potential... Type: noncoding Potential: -1.044 Folding energies... MFEI: -27.923 AMFE: -0.612 GC content %: 45.591 | |
| Tdicoccoides_TRIDC7BG042140.10 TRIDC7BG042140.10 |
Low | No | 1602 | OG0000972 | Get ORF Coding potential... Type: noncoding Potential: -1.025 Folding energies... MFEI: -30.418 AMFE: -0.592 GC content %: 51.373 | |
| Tdicoccoides_TRIDC7BG042140.11 TRIDC7BG042140.11 |
Low | No | 2616 | OG0000972 | Get ORF Coding potential... Type: noncoding Potential: -1.031 Folding energies... MFEI: -28.926 AMFE: -0.610 GC content %: 47.439 | |
| Tdicoccoides_TRIDC7BG042140.12 TRIDC7BG042140.12 |
High | No | 1059 | OG0000972 | Get ORF Coding potential... Type: noncoding Potential: -0.967 Folding energies... MFEI: -33.692 AMFE: -0.635 GC content %: 53.069 | |
| Tdicoccoides_TRIDC7BG042140.13 TRIDC7BG042140.13 |
High | No | 1112 | OG0000972 | Get ORF Coding potential... Type: noncoding Potential: -0.995 Folding energies... MFEI: -33.696 AMFE: -0.643 GC content %: 52.428 | |
| Tdicoccoides_TRIDC7BG042140.14 TRIDC7BG042140.14 |
Low | No | 1883 | OG0000972 | Get ORF Coding potential... Type: noncoding Potential: -1.035 Folding energies... MFEI: -29.841 AMFE: -0.605 GC content %: 49.336 | |
| Tdicoccoides_TRIDC7BG042140.16 TRIDC7BG042140.16 |
Low | No | 1792 | OG0367581 | Get ORF Coding potential... Type: noncoding Potential: -1.031 Folding energies... MFEI: -26.629 AMFE: -0.573 GC content %: 46.484 | |
| Tdicoccoides_TRIDC7BG042140.17 TRIDC7BG042140.17 |
Low | No | 963 | OG0000972 | Get ORF Coding potential... Type: noncoding Potential: -0.961 Folding energies... MFEI: -27.934 AMFE: -0.512 GC content %: 54.517 | |
| Tdicoccoides_TRIDC7BG042140.2 TRIDC7BG042140.2 |
Low | No | 2490 | OG0000972 | Get ORF Coding potential... Type: noncoding Potential: -1.051 Folding energies... MFEI: -29.277 AMFE: -0.622 GC content %: 47.068 | |
| Tdicoccoides_TRIDC7BG042140.3 TRIDC7BG042140.3 |
Low | No | 2074 | OG0000972 | Get ORF Coding potential... Type: noncoding Potential: -1.007 Folding energies... MFEI: -29.122 AMFE: -0.608 GC content %: 47.927 | |
| Tdicoccoides_TRIDC7BG042140.4 TRIDC7BG042140.4 |
Low | No | 3251 | OG0000972 | Get ORF Coding potential... Type: noncoding Potential: -1.051 Folding energies... MFEI: -27.410 AMFE: -0.626 GC content %: 43.771 | |
| Tdicoccoides_TRIDC7BG042140.5 TRIDC7BG042140.5 |
Low | No | 1988 | OG0000972 | Get ORF Coding potential... Type: noncoding Potential: -1.002 Folding energies... MFEI: -30.332 AMFE: -0.597 GC content %: 50.805 | |
| Tdicoccoides_TRIDC7BG042140.6 TRIDC7BG042140.6 |
High | No | 2068 | OG0000972 | Get ORF Coding potential... Type: noncoding Potential: -1.324 Folding energies... MFEI: -27.800 AMFE: -0.635 GC content %: 43.810 | |
| Tdicoccoides_TRIDC7BG042140.7 TRIDC7BG042140.7 |
Low | No | 2542 | OG0000972 | Get ORF Coding potential... Type: noncoding Potential: -1.028 Folding energies... MFEI: -28.729 AMFE: -0.605 GC content %: 47.522 | |
| Tdicoccoides_TRIDC7BG042140.8 TRIDC7BG042140.8 |
High | No | 1476 | OG0000972 | Get ORF Coding potential... Type: noncoding Potential: -1.290 Folding energies... MFEI: -30.108 AMFE: -0.614 GC content %: 49.051 | |
| Tdicoccoides_TRIDC7BG042140.9 TRIDC7BG042140.9 |
High | No | 1771 | OG0000972 | Get ORF Coding potential... Type: noncoding Potential: -1.309 Folding energies... MFEI: -29.193 AMFE: -0.624 GC content %: 46.810 |
Matches to external databases
| Transcript alias | Database | Version | Hit | Evalue |
|---|---|---|---|---|
| Tdicoccoides_TRIDC7BG042140.3 | SwissProt | 2021-04-01 | O14102 | 1.89e-4 |
| Tdicoccoides_TRIDC7BG042140.11 | SwissProt | 2021-04-01 | Q5Z7L0 | 2.28e-5 |
| Tdicoccoides_TRIDC7BG042140.4 | SwissProt | 2021-04-01 | Q5Z7L0 | 3.0e-5 |
| Tdicoccoides_TRIDC7BG042140.14 | SwissProt | 2021-04-01 | Q5Z7L0 | 1.45e-5 |
| Tdicoccoides_TRIDC7BG042140.5 | SwissProt | 2021-04-01 | Q5Z7L0 | 1.57e-5 |
| Tdicoccoides_TRIDC7BG042140.16 | SwissProt | 2021-04-01 | Q5Z7L0 | 4.5e-5 |
| Tdicoccoides_TRIDC7BG042140.7 | SwissProt | 2021-04-01 | Q5Z7L0 | 2.2e-5 |
| Tdicoccoides_TRIDC7BG042140.17 | SwissProt | 2021-04-01 | Q5Z7L0 | 4.14e-6 |
| Tdicoccoides_TRIDC7BG042140.1 | SwissProt | 2021-04-01 | Q5Z7L0 | 2.72e-5 |
| Tdicoccoides_TRIDC7BG042140.2 | SwissProt | 2021-04-01 | Q5Z7L0 | 2.14e-5 |
... further results
Gene models