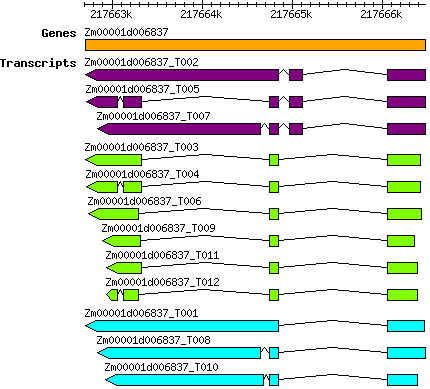
Zmays Zm00001d006837
From GreeNC
Gene information
| Gene alias / name | Chrom. | Start | End | Assembly | Species | Details
|
|---|---|---|---|---|---|---|
| Zmays_Zm00001d006837 [ Zm00001d006837] |
2 | 217662701 | 217666477 | v4 | Zea mays (Phytozome 13) | Coding? Yes |
Transcript features
This table shows how a lncRNA has been classified (high/low confidence) or whether it has been classified as pre-miRNA (go to FAQ for classification criteria). Besides, it displays the lncRNA length and sequence as well as information about the ORF, the coding potential calculated by the Coding Potential Calculator (CPC), and the folding energies calculated by RNAfold.
| Transcript alias / name | Confidence | Pre-miRNA | Length | Orthologous group | Sequence | Details
|
|---|---|---|---|---|---|---|
| Zmays_Zm00001d006837_T001 Zm00001d006837_T001 |
Low | No | 2553 | [1] | Get ORF Coding potential... Type: noncoding Potential: -0.380 Folding energies... MFEI: -27.959 AMFE: -0.690 GC content %: 40.541 | |
| Zmays_Zm00001d006837_T003 Zm00001d006837_T003 |
High | No | 1088 | [2] | Get ORF Coding potential... Type: noncoding Potential: -1.248 Folding energies... MFEI: -30.147 AMFE: -0.651 GC content %: 46.324 | |
| Zmays_Zm00001d006837_T004 Zm00001d006837_T004 |
High | No | 1003 | [3] | Get ORF Coding potential... Type: noncoding Potential: -1.237 Folding energies... MFEI: -29.322 AMFE: -0.634 GC content %: 46.261 | |
| Zmays_Zm00001d006837_T006 Zm00001d006837_T006 |
High | No | 1037 | [4] | Get ORF Coding potential... Type: noncoding Potential: -1.242 Folding energies... MFEI: -30.154 AMFE: -0.646 GC content %: 46.673 | |
| Zmays_Zm00001d006837_T008 Zm00001d006837_T008 |
Low | No | 2339 | [5] | Get ORF Coding potential... Type: noncoding Potential: -0.087 Folding energies... MFEI: -28.123 AMFE: -0.684 GC content %: 41.129 | |
| Zmays_Zm00001d006837_T009 Zm00001d006837_T009 |
High | No | 815 | [6] | Get ORF Coding potential... Type: noncoding Potential: -1.208 Folding energies... MFEI: -30.123 AMFE: -0.643 GC content %: 46.871 | |
| Zmays_Zm00001d006837_T010 Zm00001d006837_T010 |
Low | No | 2183 | [7] | Get ORF Coding potential... Type: noncoding Potential: -0.319 Folding energies... MFEI: -27.595 AMFE: -0.674 GC content %: 40.953 | |
| Zmays_Zm00001d006837_T011 Zm00001d006837_T011 |
High | No | 824 | [8] | Get ORF Coding potential... Type: noncoding Potential: -1.210 Folding energies... MFEI: -31.141 AMFE: -0.630 GC content %: 49.393 | |
| Zmays_Zm00001d006837_T012 Zm00001d006837_T012 |
High | No | 714 | [9] | Get ORF Coding potential... Type: noncoding Potential: -1.186 Folding energies... MFEI: -30.784 AMFE: -0.617 GC content %: 49.860 |
Matches to external databases
| Transcript alias | Database | Version | Hit | Evalue |
|---|---|---|---|---|
| Zmays_Zm00001d006837_T001 | SwissProt | 2021-04-01 | Q9LJE5 | 9.41e-17 |
| Zmays_Zm00001d006837_T008 | SwissProt | 2021-04-01 | Q9LJE5 | 4.74e-24 |
| Zmays_Zm00001d006837_T010 | SwissProt | 2021-04-01 | Q9LJE5 | 4.13e-18 |
Gene models