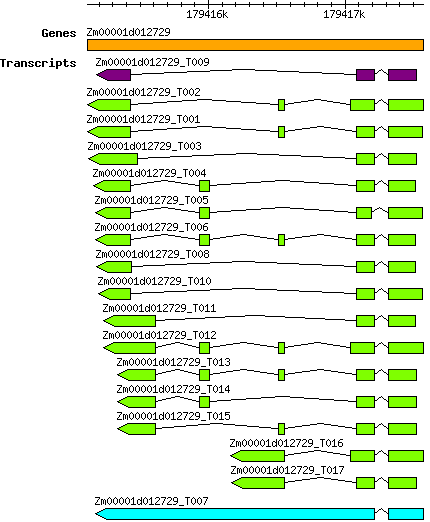
Zmays Zm00001d012729
From GreeNC
Gene information
| Gene alias / name | Chrom. | Start | End | Assembly | Species | Details
|
|---|---|---|---|---|---|---|
| Zmays_Zm00001d012729 [ Zm00001d012729] |
8 | 179415114 | 179417574 | v4 | Zea mays (Phytozome 13) | Coding? Yes |
Transcript features
This table shows how a lncRNA has been classified (high/low confidence) or whether it has been classified as pre-miRNA (go to FAQ for classification criteria). Besides, it displays the lncRNA length and sequence as well as information about the ORF, the coding potential calculated by the Coding Potential Calculator (CPC), and the folding energies calculated by RNAfold.
| Transcript alias / name | Confidence | Pre-miRNA | Length | Orthologous group | Sequence | Details
|
|---|---|---|---|---|---|---|
| Zmays_Zm00001d012729_T001 Zm00001d012729_T001 |
High | No | 728 | [1] | Get ORF Coding potential... Type: noncoding Potential: -0.984 Folding energies... MFEI: -34.725 AMFE: -0.650 GC content %: 53.434 | |
| Zmays_Zm00001d012729_T002 Zm00001d012729_T002 |
High | No | 780 | [2] | Get ORF Coding potential... Type: noncoding Potential: -0.975 Folding energies... MFEI: -34.603 AMFE: -0.646 GC content %: 53.590 | |
| Zmays_Zm00001d012729_T003 Zm00001d012729_T003 |
High | No | 687 | [3] | Get ORF Coding potential... Type: noncoding Potential: -0.969 Folding energies... MFEI: -33.886 AMFE: -0.650 GC content %: 52.111 | |
| Zmays_Zm00001d012729_T004 Zm00001d012729_T004 |
High | No | 665 | [4] | Get ORF Coding potential... Type: noncoding Potential: -1.016 Folding energies... MFEI: -36.707 AMFE: -0.682 GC content %: 53.835 | |
| Zmays_Zm00001d012729_T005 Zm00001d012729_T005 |
High | No | 680 | [5] | Get ORF Coding potential... Type: noncoding Potential: -1.075 Folding energies... MFEI: -35.265 AMFE: -0.648 GC content %: 54.412 | |
| Zmays_Zm00001d012729_T006 Zm00001d012729_T006 |
High | No | 691 | [6] | Get ORF Coding potential... Type: noncoding Potential: -1.020 Folding energies... MFEI: -34.978 AMFE: -0.653 GC content %: 53.546 | |
| Zmays_Zm00001d012729_T007 Zm00001d012729_T007 |
Low | No | 2294 | [7] | Get ORF Coding potential... Type: noncoding Potential: -1.103 Folding energies... MFEI: -31.059 AMFE: -0.720 GC content %: 43.156 | |
| Zmays_Zm00001d012729_T008 Zm00001d012729_T008 |
High | No | 581 | [8] | Get ORF Coding potential... Type: noncoding Potential: -0.999 Folding energies... MFEI: -36.024 AMFE: -0.654 GC content %: 55.077 | |
| Zmays_Zm00001d012729_T010 Zm00001d012729_T010 |
High | No | 607 | [9] | Get ORF Coding potential... Type: noncoding Potential: -0.903 Folding energies... MFEI: -36.277 AMFE: -0.642 GC content %: 56.507 | |
| Zmays_Zm00001d012729_T011 Zm00001d012729_T011 |
High | No | 709 | [10] | Get ORF Coding potential... Type: noncoding Potential: -0.947 Folding energies... MFEI: -33.399 AMFE: -0.637 GC content %: 52.468 | |
| Zmays_Zm00001d012729_T012 Zm00001d012729_T012 |
High | No | 922 | [11] | Get ORF Coding potential... Type: noncoding Potential: -0.995 Folding energies... MFEI: -34.078 AMFE: -0.652 GC content %: 52.278 | |
| Zmays_Zm00001d012729_T013 Zm00001d012729_T013 |
High | No | 719 | [12] | Get ORF Coding potential... Type: noncoding Potential: -1.031 Folding energies... MFEI: -33.741 AMFE: -0.630 GC content %: 53.547 | |
| Zmays_Zm00001d012729_T014 Zm00001d012729_T014 |
High | No | 677 | [13] | Get ORF Coding potential... Type: noncoding Potential: -1.024 Folding energies... MFEI: -34.357 AMFE: -0.636 GC content %: 54.062 | |
| Zmays_Zm00001d012729_T015 Zm00001d012729_T015 |
High | No | 642 | [14] | Get ORF Coding potential... Type: noncoding Potential: -1.017 Folding energies... MFEI: -34.159 AMFE: -0.627 GC content %: 54.517 | |
| Zmays_Zm00001d012729_T016 Zm00001d012729_T016 |
High | No | 820 | [15] | Get ORF Coding potential... Type: noncoding Potential: -0.981 Folding energies... MFEI: -34.171 AMFE: -0.637 GC content %: 53.659 | |
| Zmays_Zm00001d012729_T017 Zm00001d012729_T017 |
High | No | 719 | [16] | Get ORF Coding potential... Type: noncoding Potential: -1.031 Folding energies... MFEI: -34.103 AMFE: -0.640 GC content %: 53.268 |
Matches to external databases
| Transcript alias | Database | Version | Hit | Evalue |
|---|---|---|---|---|
| Zmays_Zm00001d012729_T007 | RepBase (RepeatMasker) | 0000-00-00 | RC/Helitron |
Gene models