Zmays Zm00001d027271
From GreeNC
Gene information
| Gene alias / name | Chrom. | Start | End | Assembly | Species | Details
|
|---|---|---|---|---|---|---|
| Zmays_Zm00001d027271 [ Zm00001d027271] |
1 | 1067813 | 1073269 | v4 | Zea mays (Phytozome 13) | Coding? Yes |
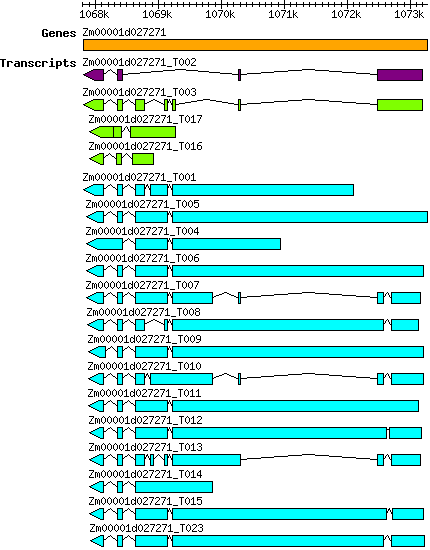
Transcript features
This table shows how a lncRNA has been classified (high/low confidence) or whether it has been classified as pre-miRNA (go to FAQ for classification criteria). Besides, it displays the lncRNA length and sequence as well as information about the ORF, the coding potential calculated by the Coding Potential Calculator (CPC), and the folding energies calculated by RNAfold.
| Transcript alias / name | Confidence | Pre-miRNA | Length | Orthologous group | Sequence | Details
|
|---|---|---|---|---|---|---|
| Zmays_Zm00001d027271_T001 Zm00001d027271_T001 |
Low | No | 3700 | [1] | Get ORF Coding potential... Type: noncoding Potential: -0.583 Folding energies... MFEI: -25.162 AMFE: -0.686 GC content %: 36.703 | |
| Zmays_Zm00001d027271_T003 Zm00001d027271_T003 |
High | No | 1408 | [2] | Get ORF Coding potential... Type: noncoding Potential: -1.164 Folding energies... MFEI: -31.974 AMFE: -0.610 GC content %: 52.415 | |
| Zmays_Zm00001d027271_T004 Zm00001d027271_T004 |
Low | No | 2799 | [3] | Get ORF Coding potential... Type: noncoding Potential: -0.511 Folding energies... MFEI: -26.027 AMFE: -0.681 GC content %: 38.192 | |
| Zmays_Zm00001d027271_T005 Zm00001d027271_T005 |
Low | No | 4906 | [4] | Get ORF Coding potential... Type: noncoding Potential: -0.321 Folding energies... MFEI: -27.008 AMFE: -0.660 GC content %: 40.909 | |
| Zmays_Zm00001d027271_T006 Zm00001d027271_T006 |
Low | No | 4838 | [5] | Get ORF Coding potential... Type: noncoding Potential: -0.433 Folding energies... MFEI: -27.205 AMFE: -0.668 GC content %: 40.719 | |
| Zmays_Zm00001d027271_T007 Zm00001d027271_T007 |
Low | No | 2113 | [6] | Get ORF Coding potential... Type: noncoding Potential: -1.112 Folding energies... MFEI: -28.628 AMFE: -0.627 GC content %: 45.670 | |
| Zmays_Zm00001d027271_T008 Zm00001d027271_T008 |
Low | No | 4327 | [7] | Get ORF Coding potential... Type: noncoding Potential: -0.343 Folding energies... MFEI: -27.338 AMFE: -0.677 GC content %: 40.351 | |
| Zmays_Zm00001d027271_T009 Zm00001d027271_T009 |
Low | No | 4844 | [8] | Get ORF Coding potential... Type: noncoding Potential: -0.418 Folding energies... MFEI: -27.244 AMFE: -0.668 GC content %: 40.772 | |
| Zmays_Zm00001d027271_T010 Zm00001d027271_T010 |
Low | No | 2116 | [9] | Get ORF Coding potential... Type: noncoding Potential: -1.112 Folding energies... MFEI: -28.733 AMFE: -0.622 GC content %: 46.219 | |
| Zmays_Zm00001d027271_T011 Zm00001d027271_T011 |
Low | No | 4732 | [10] | Get ORF Coding potential... Type: noncoding Potential: -0.553 Folding energies... MFEI: -27.276 AMFE: -0.677 GC content %: 40.300 | |
| Zmays_Zm00001d027271_T012 Zm00001d027271_T012 |
Low | No | 4731 | [11] | Get ORF Coding potential... Type: noncoding Potential: -0.521 Folding energies... MFEI: -27.366 AMFE: -0.674 GC content %: 40.626 | |
| Zmays_Zm00001d027271_T013 Zm00001d027271_T013 |
Low | No | 2227 | [12] | Get ORF Coding potential... Type: noncoding Potential: -0.260 Folding energies... MFEI: -28.859 AMFE: -0.638 GC content %: 45.218 | |
| Zmays_Zm00001d027271_T014 Zm00001d027271_T014 |
Low | No | 1517 | [13] | Get ORF Coding potential... Type: noncoding Potential: -1.254 Folding energies... MFEI: -25.188 AMFE: -0.652 GC content %: 38.629 | |
| Zmays_Zm00001d027271_T015 Zm00001d027271_T015 |
Low | No | 4705 | [14] | Get ORF Coding potential... Type: noncoding Potential: -0.416 Folding energies... MFEI: -27.290 AMFE: -0.672 GC content %: 40.638 | |
| Zmays_Zm00001d027271_T016 Zm00001d027271_T016 |
High | No | 651 | [15] | Get ORF Coding potential... Type: noncoding Potential: -1.130 Folding energies... MFEI: -25.084 AMFE: -0.603 GC content %: 41.628 | |
| Zmays_Zm00001d027271_T017 Zm00001d027271_T017 |
High | No | 1213 | [16] | Get ORF Coding potential... Type: noncoding Potential: -1.201 Folding energies... MFEI: -25.911 AMFE: -0.652 GC content %: 39.736 | |
| Zmays_Zm00001d027271_T023 Zm00001d027271_T023 |
Low | No | 4697 | [17] | Get ORF Coding potential... Type: noncoding Potential: -0.302 Folding energies... MFEI: -27.049 AMFE: -0.665 GC content %: 40.664 |
Matches to external databases
| Transcript alias | Database | Version | Hit | Evalue |
|---|---|---|---|---|
| Zmays_Zm00001d027271_T006 | SwissProt | 2021-04-01 | Q9XES0 | 1.55e-11 |
| Zmays_Zm00001d027271_T012 | SwissProt | 2021-04-01 | Q9XES0 | 1.5e-11 |
| Zmays_Zm00001d027271_T007 | SwissProt | 2021-04-01 | Q9XES0 | 3.82e-9 |
| Zmays_Zm00001d027271_T013 | SwissProt | 2021-04-01 | Q9XES0 | 5.46e-12 |
| Zmays_Zm00001d027271_T008 | SwissProt | 2021-04-01 | Q9XES0 | 1.35e-11 |
| Zmays_Zm00001d027271_T001 | SwissProt | 2021-04-01 | Q9XES0 | 1.11e-11 |
| Zmays_Zm00001d027271_T014 | SwissProt | 2021-04-01 | Q9XES0 | 1.09e-7 |
| Zmays_Zm00001d027271_T009 | SwissProt | 2021-04-01 | Q9XES0 | 1.55e-11 |
| Zmays_Zm00001d027271_T004 | SwissProt | 2021-04-01 | Q9XES0 | 7.64e-12 |
| Zmays_Zm00001d027271_T015 | SwissProt | 2021-04-01 | Q9XES0 | 1.49e-11 |
... further results
Gene models