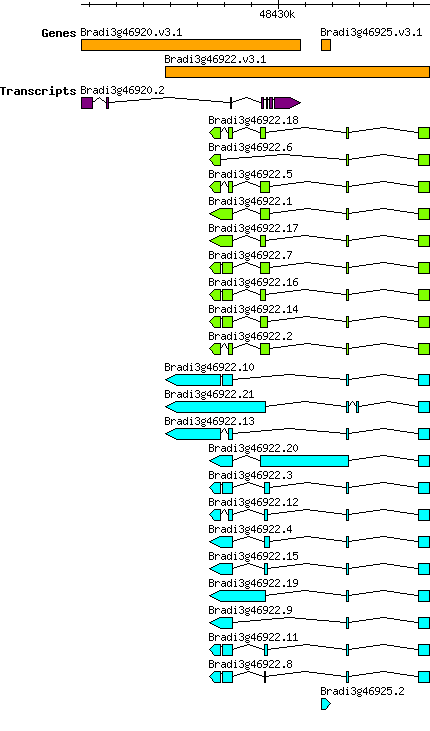
Gene
Bdistachyon Bradi3g46922
From GreeNC
Gene information
| Gene alias / name | Chrom. | Start | End | Source | Assembly | Species | Details
|
|---|---|---|---|---|---|---|---|
| Bdistachyon_Bradi3g46922 Bradi3g46922 |
Bd3 | 48425813 | 48435565 | Phytozome | v3.1 | Brachypodium_distachyon | Coding? No |
Transcript features
This table shows how a lncRNA has been classified (high/low confidence) or whether it has been classified as pre-miRNA (go to FAQ for classification criteria). Besides, it displays the lncRNA length and sequence as well as information about the ORF, the coding potential calculated by the Coding Potential Calculator (CPC), and the folding energies calculated by RNAfold.
| Transcript alias / name | Confidence | Pre-miRNA | Length | Sequence | Details
|
|---|---|---|---|---|---|
| Bdistachyon_Bradi3g46922.1 Bradi3g46922.1 PAC:32816308 |
High | No | 1616 | Get ORF Coding potential... Type: noncoding Potential: -1.208 Folding energies... MFEI: -28.131 AMFE: -0.613 GC content %: 45.854 | |
| Bdistachyon_Bradi3g46922.10 Bradi3g46922.10 PAC:32816322 |
Low | Yes | 2834 | Get ORF Coding potential... Type: noncoding Potential: -1.114 Folding energies... MFEI: -28.052 AMFE: -0.613 GC content %: 45.730 | |
| Bdistachyon_Bradi3g46922.11 Bradi3g46922.11 PAC:32816316 |
Low | Yes | 1316 | Get ORF Coding potential... Type: noncoding Potential: -1.162 Folding energies... MFEI: -28.997 AMFE: -0.617 GC content %: 46.960 | |
| Bdistachyon_Bradi3g46922.12 Bradi3g46922.12 PAC:32816317 |
Low | Yes | 1077 | Get ORF Coding potential... Type: noncoding Potential: -1.127 Folding energies... MFEI: -29.861 AMFE: -0.606 GC content %: 49.304 | |
| Bdistachyon_Bradi3g46922.13 Bradi3g46922.13 PAC:32816318 |
Low | Yes | 2592 | Get ORF Coding potential... Type: noncoding Potential: -1.090 Folding energies... MFEI: -28.302 AMFE: -0.608 GC content %: 46.566 | |
| Bdistachyon_Bradi3g46922.14 Bradi3g46922.14 PAC:32816319 |
High | No | 1463 | Get ORF Coding potential... Type: noncoding Potential: -1.249 Folding energies... MFEI: -28.503 AMFE: -0.610 GC content %: 46.753 | |
| Bdistachyon_Bradi3g46922.15 Bradi3g46922.15 PAC:32816320 |
Low | Yes | 1399 | Get ORF Coding potential... Type: noncoding Potential: -1.170 Folding energies... MFEI: -29.257 AMFE: -0.627 GC content %: 46.676 | |
| Bdistachyon_Bradi3g46922.16 Bradi3g46922.16 PAC:32816326 |
High | No | 1370 | Get ORF Coding potential... Type: noncoding Potential: -1.244 Folding energies... MFEI: -29.102 AMFE: -0.615 GC content %: 47.299 | |
| Bdistachyon_Bradi3g46922.17 Bradi3g46922.17 PAC:32816323 |
High | No | 1451 | Get ORF Coding potential... Type: noncoding Potential: -1.248 Folding energies... MFEI: -29.387 AMFE: -0.626 GC content %: 46.933 | |
| Bdistachyon_Bradi3g46922.18 Bradi3g46922.18 PAC:32816324 |
High | No | 1128 | Get ORF Coding potential... Type: noncoding Potential: -1.230 Folding energies... MFEI: -30.027 AMFE: -0.606 GC content %: 49.557 | |
| Bdistachyon_Bradi3g46922.19 Bradi3g46922.19 PAC:32816325 |
Low | Yes | 2514 | Get ORF Coding potential... Type: noncoding Potential: -1.161 Folding energies... MFEI: -29.372 AMFE: -0.699 GC content %: 42.005 | |
| Bdistachyon_Bradi3g46922.2 Bradi3g46922.2 PAC:32816309 |
High | No | 1294 | Get ORF Coding potential... Type: noncoding Potential: -1.185 Folding energies... MFEI: -28.354 AMFE: -0.593 GC content %: 47.836 | |
| Bdistachyon_Bradi3g46922.20 Bradi3g46922.20 PAC:32816327 |
Low | No | 4469 | Get ORF Coding potential... Type: noncoding Potential: -1.068 Folding energies... MFEI: -26.044 AMFE: -0.619 GC content %: 42.068 | |
| Bdistachyon_Bradi3g46922.21 Bradi3g46922.21 PAC:32816328 |
Low | Yes | 4196 | Get ORF Coding potential... Type: noncoding Potential: -1.188 Folding energies... MFEI: -28.384 AMFE: -0.659 GC content %: 43.065 | |
| Bdistachyon_Bradi3g46922.3 Bradi3g46922.3 PAC:32816310 |
Low | Yes | 1386 | Get ORF Coding potential... Type: noncoding Potential: -0.876 Folding energies... MFEI: -28.102 AMFE: -0.609 GC content %: 46.176 | |
| Bdistachyon_Bradi3g46922.4 Bradi3g46922.4 PAC:32816311 |
Low | Yes | 1469 | Get ORF Coding potential... Type: noncoding Potential: -0.887 Folding energies... MFEI: -28.400 AMFE: -0.618 GC content %: 45.950 | |
| Bdistachyon_Bradi3g46922.5 Bradi3g46922.5 PAC:32816314 |
High | No | 1291 | Get ORF Coding potential... Type: noncoding Potential: -1.185 Folding energies... MFEI: -28.420 AMFE: -0.594 GC content %: 47.870 | |
| Bdistachyon_Bradi3g46922.6 Bradi3g46922.6 PAC:32816312 |
High | No | 856 | Get ORF Coding potential... Type: noncoding Potential: -1.140 Folding energies... MFEI: -27.979 AMFE: -0.547 GC content %: 51.168 | |
| Bdistachyon_Bradi3g46922.7 Bradi3g46922.7 PAC:32816313 |
High | No | 1533 | Get ORF Coding potential... Type: noncoding Potential: -1.203 Folding energies... MFEI: -27.847 AMFE: -0.604 GC content %: 46.119 | |
| Bdistachyon_Bradi3g46922.8 Bradi3g46922.8 PAC:32816315 |
Low | Yes | 1220 | Get ORF Coding potential... Type: noncoding Potential: -1.236 Folding energies... MFEI: -29.951 AMFE: -0.630 GC content %: 47.541 | |
| Bdistachyon_Bradi3g46922.9 Bradi3g46922.9 PAC:32816321 |
Low | Yes | 1297 | Get ORF Coding potential... Type: noncoding Potential: -1.020 Folding energies... MFEI: -30.162 AMFE: -0.638 GC content %: 47.263 |
Matches to external databases
| Transcript alias | Database | Version | Hit | Evalue |
|---|---|---|---|---|
| Bdistachyon_Bradi3g46922.21 | Rfam | 11.0 | MIR1122 | 3.5e-16 |
| Bdistachyon_Bradi3g46922.11 | Rfam | 11.0 | MIR444 | 9.7e-25 |
| Bdistachyon_Bradi3g46922.20 | RepBase (RepeatMasker) | 2014-01-31 | L1-8_SBi | |
| Bdistachyon_Bradi3g46922.3 | Rfam | 11.0 | MIR444 | 0.0065 |
| Bdistachyon_Bradi3g46922.12 | Rfam | 11.0 | MIR444 | 8.0e-25 |
| Bdistachyon_Bradi3g46922.21 | RepBase (RepeatMasker) | 2014-01-31 | TREP216 | |
| Bdistachyon_Bradi3g46922.4 | Rfam | 11.0 | MIR444 | 0.0069 |
| Bdistachyon_Bradi3g46922.13 | Rfam | 11.0 | MIR444 | 4.4e-29 |
| Bdistachyon_Bradi3g46922.8 | Rfam | 11.0 | MIR444 | 7.1e-31 |
| Bdistachyon_Bradi3g46922.15 | Rfam | 11.0 | MIR444 | 1.0e-24 |
... further results
Gene models