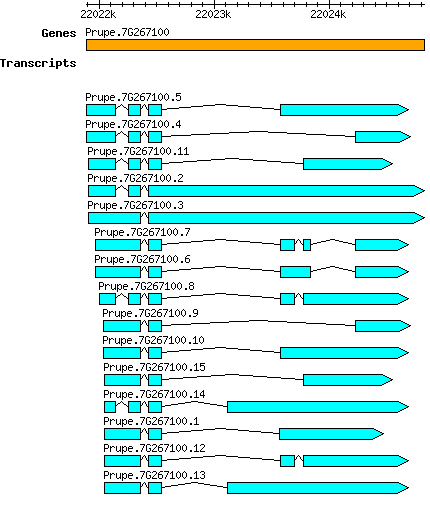
Gene
Ppersica Prupe.7G267100
From GreeNC
Gene information
| Gene alias / name | Chrom. | Start | End | Source | Assembly | Species | Details
|
|---|---|---|---|---|---|---|---|
| Ppersica_Prupe.7G267100 Prupe.7G267100 |
Pp07 | 22021886 | 22024828 | Phytozome | v2.1 | Prunus_persica | Coding? No |
Transcript features
This table shows how a lncRNA has been classified (high/low confidence) or whether it has been classified as pre-miRNA (go to FAQ for classification criteria). Besides, it displays the lncRNA length and sequence as well as information about the ORF, the coding potential calculated by the Coding Potential Calculator (CPC), and the folding energies calculated by RNAfold.
| Transcript alias / name | Confidence | Pre-miRNA | Length | Sequence | Details
|
|---|---|---|---|---|---|
| Ppersica_Prupe.7G267100.1 Prupe.7G267100.1 PAC:32101081 |
Low | No | 1336 | Get ORF Coding potential... Type: noncoding Potential: -1.311 Folding energies... MFEI: -25.516 AMFE: -0.654 GC content %: 38.997 | |
| Ppersica_Prupe.7G267100.10 Prupe.7G267100.10 PAC:32101090 |
Low | No | 1543 | Get ORF Coding potential... Type: noncoding Potential: -1.317 Folding energies... MFEI: -25.800 AMFE: -0.655 GC content %: 39.404 | |
| Ppersica_Prupe.7G267100.11 Prupe.7G267100.11 PAC:32101089 |
Low | No | 1235 | Get ORF Coding potential... Type: noncoding Potential: -1.307 Folding energies... MFEI: -24.964 AMFE: -0.650 GC content %: 38.381 | |
| Ppersica_Prupe.7G267100.12 Prupe.7G267100.12 PAC:32101091 |
Low | No | 1468 | Get ORF Coding potential... Type: noncoding Potential: -1.245 Folding energies... MFEI: -25.770 AMFE: -0.649 GC content %: 39.714 | |
| Ppersica_Prupe.7G267100.13 Prupe.7G267100.13 PAC:32101082 |
Low | No | 2001 | Get ORF Coding potential... Type: noncoding Potential: -1.327 Folding energies... MFEI: -24.378 AMFE: -0.638 GC content %: 38.181 | |
| Ppersica_Prupe.7G267100.14 Prupe.7G267100.14 PAC:32101083 |
Low | No | 1888 | Get ORF Coding potential... Type: noncoding Potential: -1.325 Folding energies... MFEI: -24.555 AMFE: -0.643 GC content %: 38.189 | |
| Ppersica_Prupe.7G267100.15 Prupe.7G267100.15 PAC:32101088 |
Low | No | 1209 | Get ORF Coding potential... Type: noncoding Potential: -1.306 Folding energies... MFEI: -23.921 AMFE: -0.633 GC content %: 37.800 | |
| Ppersica_Prupe.7G267100.2 Prupe.7G267100.2 PAC:32101084 |
Low | No | 2745 | Get ORF Coding potential... Type: noncoding Potential: -1.159 Folding energies... MFEI: -24.863 AMFE: -0.658 GC content %: 37.814 | |
| Ppersica_Prupe.7G267100.3 Prupe.7G267100.3 PAC:32101085 |
Low | No | 2857 | Get ORF Coding potential... Type: noncoding Potential: -1.161 Folding energies... MFEI: -24.886 AMFE: -0.658 GC content %: 37.802 | |
| Ppersica_Prupe.7G267100.4 Prupe.7G267100.4 PAC:32101087 |
Low | No | 955 | Get ORF Coding potential... Type: noncoding Potential: -1.293 Folding energies... MFEI: -27.916 AMFE: -0.663 GC content %: 42.094 | |
| Ppersica_Prupe.7G267100.5 Prupe.7G267100.5 PAC:32101093 |
Low | No | 1584 | Get ORF Coding potential... Type: noncoding Potential: -1.318 Folding energies... MFEI: -26.673 AMFE: -0.665 GC content %: 40.088 | |
| Ppersica_Prupe.7G267100.6 Prupe.7G267100.6 PAC:32101094 |
Low | No | 1224 | Get ORF Coding potential... Type: noncoding Potential: -1.307 Folding energies... MFEI: -27.827 AMFE: -0.663 GC content %: 41.993 | |
| Ppersica_Prupe.7G267100.7 Prupe.7G267100.7 PAC:32101095 |
Low | No | 1150 | Get ORF Coding potential... Type: noncoding Potential: -1.177 Folding energies... MFEI: -27.974 AMFE: -0.658 GC content %: 42.522 | |
| Ppersica_Prupe.7G267100.8 Prupe.7G267100.8 PAC:32101092 |
Low | No | 1396 | Get ORF Coding potential... Type: noncoding Potential: -1.240 Folding energies... MFEI: -26.590 AMFE: -0.657 GC content %: 40.473 | |
| Ppersica_Prupe.7G267100.9 Prupe.7G267100.9 PAC:32101086 |
Low | No | 913 | Get ORF Coding potential... Type: noncoding Potential: -1.290 Folding energies... MFEI: -26.473 AMFE: -0.645 GC content %: 41.073 |
Matches to external databases
| Transcript alias | Database | Version | Hit | Evalue |
|---|---|---|---|---|
| Ppersica_Prupe.7G267100.7 | RepBase (RepeatMasker) | 2014-01-31 | MuDR-9_Mad | |
| Ppersica_Prupe.7G267100.2 | RepBase (RepeatMasker) | 2014-01-31 | MuDR-9_Mad | |
| Ppersica_Prupe.7G267100.11 | RepBase (RepeatMasker) | 2014-01-31 | MuDR-9_Mad | |
| Ppersica_Prupe.7G267100.8 | RepBase (RepeatMasker) | 2014-01-31 | MuDR-9_Mad | |
| Ppersica_Prupe.7G267100.3 | RepBase (RepeatMasker) | 2014-01-31 | MuDR-9_Mad | |
| Ppersica_Prupe.7G267100.12 | RepBase (RepeatMasker) | 2014-01-31 | MuDR-9_Mad | |
| Ppersica_Prupe.7G267100.9 | RepBase (RepeatMasker) | 2014-01-31 | MuDR-9_Mad | |
| Ppersica_Prupe.7G267100.4 | RepBase (RepeatMasker) | 2014-01-31 | MuDR-9_Mad | |
| Ppersica_Prupe.7G267100.13 | RepBase (RepeatMasker) | 2014-01-31 | MuDR-9_Mad | |
| Ppersica_Prupe.7G267100.5 | RepBase (RepeatMasker) | 2014-01-31 | MuDR-9_Mad |
... further results
Gene models