Gene
Taestivum Traes 1AL 4273DEC3A
From GreeNC
Gene information
| Gene alias / name | Chrom. | Start | End | Source | Assembly | Species | Details
|
|---|---|---|---|---|---|---|---|
| Taestivum_Traes_1AL_4273DEC3A Traes_1AL_4273DEC3A |
ta_iwgsc_1al_v2_3876672 | 733 | 7496 | Phytozome | v2.2 | Triticum_aestivum | Coding? No |
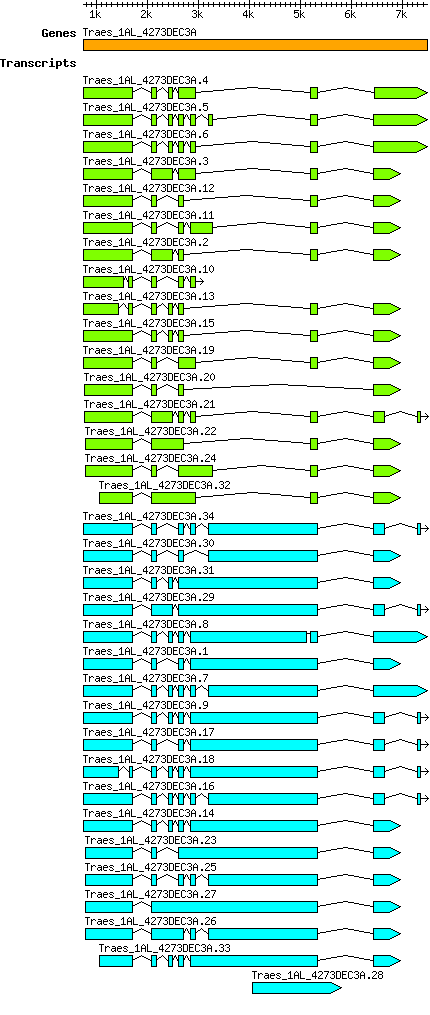
Transcript features
This table shows how a lncRNA has been classified (high/low confidence) or whether it has been classified as pre-miRNA (go to FAQ for classification criteria). Besides, it displays the lncRNA length and sequence as well as information about the ORF, the coding potential calculated by the Coding Potential Calculator (CPC), and the folding energies calculated by RNAfold.
| Transcript alias / name | Confidence | Pre-miRNA | Length | Sequence | Details
|
|---|---|---|---|---|---|
| Taestivum_Traes_1AL_4273DEC3A.1 Traes_1AL_4273DEC3A.1 PAC:32009072 |
Low | No | 4190 | Get ORF Coding potential... Type: noncoding Potential: -0.384 Folding energies... MFEI: -32.055 AMFE: -0.701 GC content %: 45.728 | |
| Taestivum_Traes_1AL_4273DEC3A.10 Traes_1AL_4273DEC3A.10 PAC:32009077 |
High | No | 1165 | Get ORF Coding potential... Type: noncoding Potential: -0.242 Folding energies... MFEI: -37.991 AMFE: -0.688 GC content %: 55.193 | |
| Taestivum_Traes_1AL_4273DEC3A.11 Traes_1AL_4273DEC3A.11 PAC:32009075 |
High | No | 2278 | Get ORF Coding potential... Type: noncoding Potential: -0.326 Folding energies... MFEI: -34.552 AMFE: -0.705 GC content %: 49.034 | |
| Taestivum_Traes_1AL_4273DEC3A.12 Traes_1AL_4273DEC3A.12 PAC:32009073 |
High | No | 1829 | Get ORF Coding potential... Type: noncoding Potential: -0.300 Folding energies... MFEI: -36.085 AMFE: -0.699 GC content %: 51.613 | |
| Taestivum_Traes_1AL_4273DEC3A.13 Traes_1AL_4273DEC3A.13 PAC:32009078 |
High | No | 1716 | Get ORF Coding potential... Type: noncoding Potential: -0.300 Folding energies... MFEI: -36.620 AMFE: -0.690 GC content %: 53.089 | |
| Taestivum_Traes_1AL_4273DEC3A.14 Traes_1AL_4273DEC3A.14 PAC:32009083 |
Low | No | 4268 | Get ORF Coding potential... Type: noncoding Potential: -0.385 Folding energies... MFEI: -32.078 AMFE: -0.702 GC content %: 45.689 | |
| Taestivum_Traes_1AL_4273DEC3A.15 Traes_1AL_4273DEC3A.15 PAC:32009084 |
High | No | 1907 | Get ORF Coding potential... Type: noncoding Potential: -0.305 Folding energies... MFEI: -35.931 AMFE: -0.701 GC content %: 51.285 | |
| Taestivum_Traes_1AL_4273DEC3A.16 Traes_1AL_4273DEC3A.16 PAC:32009082 |
Low | No | 3739 | Get ORF Coding potential... Type: noncoding Potential: -0.375 Folding energies... MFEI: -32.795 AMFE: -0.703 GC content %: 46.617 | |
| Taestivum_Traes_1AL_4273DEC3A.17 Traes_1AL_4273DEC3A.17 PAC:32009080 |
Low | No | 3921 | Get ORF Coding potential... Type: noncoding Potential: -0.379 Folding energies... MFEI: -32.339 AMFE: -0.704 GC content %: 45.907 | |
| Taestivum_Traes_1AL_4273DEC3A.18 Traes_1AL_4273DEC3A.18 PAC:32009081 |
Low | No | 3799 | Get ORF Coding potential... Type: noncoding Potential: -0.376 Folding energies... MFEI: -32.530 AMFE: -0.701 GC content %: 46.433 | |
| Taestivum_Traes_1AL_4273DEC3A.19 Traes_1AL_4273DEC3A.19 PAC:32009085 |
High | No | 2049 | Get ORF Coding potential... Type: noncoding Potential: -0.314 Folding energies... MFEI: -35.959 AMFE: -0.710 GC content %: 50.659 | |
| Taestivum_Traes_1AL_4273DEC3A.2 Traes_1AL_4273DEC3A.2 PAC:32009076 |
High | No | 2135 | Get ORF Coding potential... Type: noncoding Potential: -0.319 Folding energies... MFEI: -35.382 AMFE: -0.712 GC content %: 49.696 | |
| Taestivum_Traes_1AL_4273DEC3A.20 Traes_1AL_4273DEC3A.20 PAC:32009086 |
High | No | 1668 | Get ORF Coding potential... Type: noncoding Potential: -0.288 Folding energies... MFEI: -36.037 AMFE: -0.697 GC content %: 51.679 | |
| Taestivum_Traes_1AL_4273DEC3A.21 Traes_1AL_4273DEC3A.21 PAC:32009087 |
High | No | 1959 | Get ORF Coding potential... Type: noncoding Potential: -0.309 Folding energies... MFEI: -36.192 AMFE: -0.722 GC content %: 50.128 | |
| Taestivum_Traes_1AL_4273DEC3A.22 Traes_1AL_4273DEC3A.22 PAC:32009088 |
High | No | 2210 | Get ORF Coding potential... Type: noncoding Potential: -0.332 Folding energies... MFEI: -34.765 AMFE: -0.715 GC content %: 48.597 | |
| Taestivum_Traes_1AL_4273DEC3A.23 Traes_1AL_4273DEC3A.23 PAC:32009089 |
Low | No | 4273 | Get ORF Coding potential... Type: noncoding Potential: -0.395 Folding energies... MFEI: -31.954 AMFE: -0.703 GC content %: 45.448 | |
| Taestivum_Traes_1AL_4273DEC3A.24 Traes_1AL_4273DEC3A.24 PAC:32009091 |
High | No | 2362 | Get ORF Coding potential... Type: noncoding Potential: -0.355 Folding energies... MFEI: -34.246 AMFE: -0.706 GC content %: 48.476 | |
| Taestivum_Traes_1AL_4273DEC3A.25 Traes_1AL_4273DEC3A.25 PAC:32009090 |
Low | No | 3885 | Get ORF Coding potential... Type: noncoding Potential: -0.402 Folding energies... MFEI: -32.556 AMFE: -0.704 GC content %: 46.255 | |
| Taestivum_Traes_1AL_4273DEC3A.26 Traes_1AL_4273DEC3A.26 PAC:32009093 |
Low | No | 4307 | Get ORF Coding potential... Type: noncoding Potential: -0.444 Folding energies... MFEI: -32.090 AMFE: -0.707 GC content %: 45.368 | |
| Taestivum_Traes_1AL_4273DEC3A.27 Traes_1AL_4273DEC3A.27 PAC:32009092 |
Low | No | 4696 | Get ORF Coding potential... Type: noncoding Potential: -0.450 Folding energies... MFEI: -31.618 AMFE: -0.707 GC content %: 44.740 | |
| Taestivum_Traes_1AL_4273DEC3A.28 Traes_1AL_4273DEC3A.28 PAC:32009094 |
Low | No | 1745 | Get ORF Coding potential... Type: noncoding Potential: -1.394 Folding energies... MFEI: -29.834 AMFE: -0.674 GC content %: 44.241 | |
| Taestivum_Traes_1AL_4273DEC3A.29 Traes_1AL_4273DEC3A.29 PAC:32009095 |
Low | No | 4367 | Get ORF Coding potential... Type: noncoding Potential: -0.387 Folding energies... MFEI: -32.013 AMFE: -0.706 GC content %: 45.340 | |
| Taestivum_Traes_1AL_4273DEC3A.3 Traes_1AL_4273DEC3A.3 PAC:32009071 |
High | No | 2369 | Get ORF Coding potential... Type: noncoding Potential: -0.331 Folding energies... MFEI: -35.023 AMFE: -0.714 GC content %: 49.050 | |
| Taestivum_Traes_1AL_4273DEC3A.30 Traes_1AL_4273DEC3A.30 PAC:32009096 |
Low | No | 3824 | Get ORF Coding potential... Type: noncoding Potential: -0.377 Folding energies... MFEI: -32.827 AMFE: -0.705 GC content %: 46.574 | |
| Taestivum_Traes_1AL_4273DEC3A.31 Traes_1AL_4273DEC3A.31 PAC:32009097 |
Low | No | 4403 | Get ORF Coding potential... Type: noncoding Potential: -0.388 Folding energies... MFEI: -32.044 AMFE: -0.703 GC content %: 45.605 | |
| Taestivum_Traes_1AL_4273DEC3A.32 Traes_1AL_4273DEC3A.32 PAC:32009098 |
High | No | 2174 | Get ORF Coding potential... Type: noncoding Potential: -0.994 Folding energies... MFEI: -32.374 AMFE: -0.717 GC content %: 45.124 | |
| Taestivum_Traes_1AL_4273DEC3A.33 Traes_1AL_4273DEC3A.33 PAC:32009099 |
Low | No | 3953 | Get ORF Coding potential... Type: noncoding Potential: -1.027 Folding energies... MFEI: -30.774 AMFE: -0.705 GC content %: 43.638 | |
| Taestivum_Traes_1AL_4273DEC3A.34 Traes_1AL_4273DEC3A.34 PAC:32009100 |
Low | No | 3672 | Get ORF Coding potential... Type: noncoding Potential: -0.374 Folding energies... MFEI: -32.840 AMFE: -0.703 GC content %: 46.732 | |
| Taestivum_Traes_1AL_4273DEC3A.4 Traes_1AL_4273DEC3A.4 PAC:32009067 |
High | No | 2667 | Get ORF Coding potential... Type: noncoding Potential: -0.344 Folding energies... MFEI: -32.940 AMFE: -0.696 GC content %: 47.319 | |
| Taestivum_Traes_1AL_4273DEC3A.5 Traes_1AL_4273DEC3A.5 PAC:32009069 |
High | No | 2630 | Get ORF Coding potential... Type: noncoding Potential: -0.342 Folding energies... MFEI: -32.890 AMFE: -0.694 GC content %: 47.414 | |
| Taestivum_Traes_1AL_4273DEC3A.6 Traes_1AL_4273DEC3A.6 PAC:32009070 |
High | No | 2550 | Get ORF Coding potential... Type: noncoding Potential: -0.339 Folding energies... MFEI: -32.988 AMFE: -0.695 GC content %: 47.451 | |
| Taestivum_Traes_1AL_4273DEC3A.7 Traes_1AL_4273DEC3A.7 PAC:32009074 |
Low | No | 4542 | Get ORF Coding potential... Type: noncoding Potential: -0.390 Folding energies... MFEI: -31.308 AMFE: -0.695 GC content %: 45.046 | |
| Taestivum_Traes_1AL_4273DEC3A.8 Traes_1AL_4273DEC3A.8 PAC:32009068 |
Low | No | 4722 | Get ORF Coding potential... Type: noncoding Potential: -0.393 Folding energies... MFEI: -31.076 AMFE: -0.698 GC content %: 44.494 | |
| Taestivum_Traes_1AL_4273DEC3A.9 Traes_1AL_4273DEC3A.9 PAC:32009079 |
Low | No | 4003 | Get ORF Coding potential... Type: noncoding Potential: -0.381 Folding energies... MFEI: -32.268 AMFE: -0.704 GC content %: 45.841 |
Matches to external databases
| Transcript alias | Database | Version | Hit | Evalue |
|---|---|---|---|---|
| Taestivum_Traes_1AL_4273DEC3A.28 | RepBase (RepeatMasker) | 2014-01-31 | TREP52 | |
| Taestivum_Traes_1AL_4273DEC3A.18 | RepBase (RepeatMasker) | 2014-01-31 | TREP52 | |
| Taestivum_Traes_1AL_4273DEC3A.7 | RepBase (RepeatMasker) | 2014-01-31 | TREP52 | |
| Taestivum_Traes_1AL_4273DEC3A.29 | RepBase (RepeatMasker) | 2014-01-31 | TREP52 | |
| Taestivum_Traes_1AL_4273DEC3A.23 | RepBase (RepeatMasker) | 2014-01-31 | TREP52 | |
| Taestivum_Traes_1AL_4273DEC3A.1 | RepBase (RepeatMasker) | 2014-01-31 | TREP52 | |
| Taestivum_Traes_1AL_4273DEC3A.8 | RepBase (RepeatMasker) | 2014-01-31 | TREP52 | |
| Taestivum_Traes_1AL_4273DEC3A.30 | RepBase (RepeatMasker) | 2014-01-31 | TREP52 | |
| Taestivum_Traes_1AL_4273DEC3A.25 | RepBase (RepeatMasker) | 2014-01-31 | TREP52 | |
| Taestivum_Traes_1AL_4273DEC3A.14 | RepBase (RepeatMasker) | 2014-01-31 | TREP52 |
... further results
Gene models