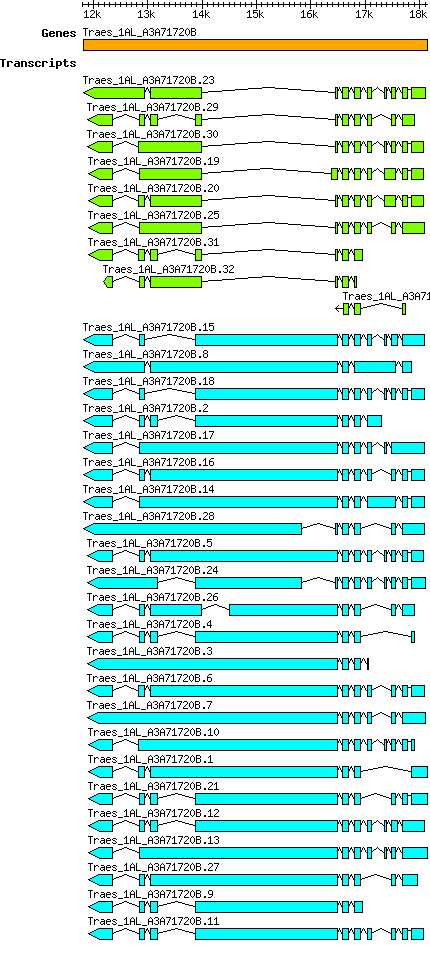
Gene
Taestivum Traes 1AL A3A71720B
From GreeNC
Gene information
| Gene alias / name | Chrom. | Start | End | Source | Assembly | Species | Details
|
|---|---|---|---|---|---|---|---|
| Taestivum_Traes_1AL_A3A71720B Traes_1AL_A3A71720B |
ta_iwgsc_1al_v2_3913351 | 11811 | 18150 | Phytozome | v2.2 | Triticum_aestivum | Coding? No |
Transcript features
This table shows how a lncRNA has been classified (high/low confidence) or whether it has been classified as pre-miRNA (go to FAQ for classification criteria). Besides, it displays the lncRNA length and sequence as well as information about the ORF, the coding potential calculated by the Coding Potential Calculator (CPC), and the folding energies calculated by RNAfold.
| Transcript alias / name | Confidence | Pre-miRNA | Length | Sequence | Details
|
|---|---|---|---|---|---|
| Taestivum_Traes_1AL_A3A71720B.1 Traes_1AL_A3A71720B.1 PAC:31932749 |
Low | No | 4538 | Get ORF Coding potential... Type: noncoding Potential: -0.683 Folding energies... MFEI: -27.512 AMFE: -0.652 GC content %: 42.177 | |
| Taestivum_Traes_1AL_A3A71720B.10 Traes_1AL_A3A71720B.10 PAC:31932751 |
Low | No | 4691 | Get ORF Coding potential... Type: noncoding Potential: -0.822 Folding energies... MFEI: -26.796 AMFE: -0.659 GC content %: 40.652 | |
| Taestivum_Traes_1AL_A3A71720B.11 Traes_1AL_A3A71720B.11 PAC:31932765 |
Low | No | 4025 | Get ORF Coding potential... Type: noncoding Potential: -0.812 Folding energies... MFEI: -28.350 AMFE: -0.656 GC content %: 43.205 | |
| Taestivum_Traes_1AL_A3A71720B.12 Traes_1AL_A3A71720B.12 PAC:31932759 |
Low | No | 4163 | Get ORF Coding potential... Type: noncoding Potential: -0.815 Folding energies... MFEI: -28.279 AMFE: -0.654 GC content %: 43.238 | |
| Taestivum_Traes_1AL_A3A71720B.13 Traes_1AL_A3A71720B.13 PAC:31932760 |
Low | No | 4988 | Get ORF Coding potential... Type: noncoding Potential: -0.825 Folding energies... MFEI: -27.556 AMFE: -0.652 GC content %: 42.281 | |
| Taestivum_Traes_1AL_A3A71720B.14 Traes_1AL_A3A71720B.14 PAC:31932762 |
Low | No | 5294 | Get ORF Coding potential... Type: noncoding Potential: -0.828 Folding energies... MFEI: -27.566 AMFE: -0.667 GC content %: 41.311 | |
| Taestivum_Traes_1AL_A3A71720B.15 Traes_1AL_A3A71720B.15 PAC:31932755 |
Low | No | 4130 | Get ORF Coding potential... Type: noncoding Potential: -0.814 Folding energies... MFEI: -28.442 AMFE: -0.657 GC content %: 43.269 | |
| Taestivum_Traes_1AL_A3A71720B.16 Traes_1AL_A3A71720B.16 PAC:31932754 |
Low | No | 4821 | Get ORF Coding potential... Type: noncoding Potential: -0.823 Folding energies... MFEI: -27.446 AMFE: -0.657 GC content %: 41.776 | |
| Taestivum_Traes_1AL_A3A71720B.17 Traes_1AL_A3A71720B.17 PAC:31932753 |
Low | No | 5150 | Get ORF Coding potential... Type: noncoding Potential: -0.826 Folding energies... MFEI: -27.337 AMFE: -0.658 GC content %: 41.534 | |
| Taestivum_Traes_1AL_A3A71720B.18 Traes_1AL_A3A71720B.18 PAC:31932766 |
Low | No | 4028 | Get ORF Coding potential... Type: noncoding Potential: -0.812 Folding energies... MFEI: -28.512 AMFE: -0.658 GC content %: 43.347 | |
| Taestivum_Traes_1AL_A3A71720B.19 Traes_1AL_A3A71720B.19 PAC:31932768 |
High | No | 2524 | Get ORF Coding potential... Type: noncoding Potential: -0.775 Folding energies... MFEI: -28.629 AMFE: -0.663 GC content %: 43.185 | |
| Taestivum_Traes_1AL_A3A71720B.2 Traes_1AL_A3A71720B.2 PAC:31932750 |
Low | No | 3891 | Get ORF Coding potential... Type: noncoding Potential: -0.810 Folding energies... MFEI: -27.299 AMFE: -0.669 GC content %: 40.786 | |
| Taestivum_Traes_1AL_A3A71720B.20 Traes_1AL_A3A71720B.20 PAC:31932767 |
High | No | 2364 | Get ORF Coding potential... Type: noncoding Potential: -0.768 Folding energies... MFEI: -29.167 AMFE: -0.671 GC content %: 43.443 | |
| Taestivum_Traes_1AL_A3A71720B.21 Traes_1AL_A3A71720B.21 PAC:31932769 |
Low | No | 4023 | Get ORF Coding potential... Type: noncoding Potential: -0.833 Folding energies... MFEI: -28.570 AMFE: -0.651 GC content %: 43.898 | |
| Taestivum_Traes_1AL_A3A71720B.22 Traes_1AL_A3A71720B.22 PAC:31932772 |
High | No | 288 | Get ORF Coding potential... Type: noncoding Potential: -0.670 Folding energies... MFEI: -34.549 AMFE: -0.634 GC content %: 54.514 | |
| Taestivum_Traes_1AL_A3A71720B.23 Traes_1AL_A3A71720B.23 PAC:31932770 |
High | No | 2869 | Get ORF Coding potential... Type: noncoding Potential: -0.787 Folding energies... MFEI: -28.348 AMFE: -0.664 GC content %: 42.663 | |
| Taestivum_Traes_1AL_A3A71720B.24 Traes_1AL_A3A71720B.24 PAC:31932771 |
Low | No | 4052 | Get ORF Coding potential... Type: noncoding Potential: -0.813 Folding energies... MFEI: -28.956 AMFE: -0.666 GC content %: 43.509 | |
| Taestivum_Traes_1AL_A3A71720B.25 Traes_1AL_A3A71720B.25 PAC:31932773 |
High | No | 2441 | Get ORF Coding potential... Type: noncoding Potential: -0.772 Folding energies... MFEI: -28.882 AMFE: -0.656 GC content %: 43.998 | |
| Taestivum_Traes_1AL_A3A71720B.26 Traes_1AL_A3A71720B.26 PAC:31932776 |
Low | No | 4016 | Get ORF Coding potential... Type: noncoding Potential: -0.833 Folding energies... MFEI: -26.414 AMFE: -0.649 GC content %: 40.687 | |
| Taestivum_Traes_1AL_A3A71720B.27 Traes_1AL_A3A71720B.27 PAC:31932774 |
Low | No | 4590 | Get ORF Coding potential... Type: noncoding Potential: -0.841 Folding energies... MFEI: -27.115 AMFE: -0.659 GC content %: 41.176 | |
| Taestivum_Traes_1AL_A3A71720B.28 Traes_1AL_A3A71720B.28 PAC:31932775 |
Low | No | 4791 | Get ORF Coding potential... Type: noncoding Potential: -0.844 Folding energies... MFEI: -27.863 AMFE: -0.664 GC content %: 41.975 | |
| Taestivum_Traes_1AL_A3A71720B.29 Traes_1AL_A3A71720B.29 PAC:31932777 |
High | No | 1457 | Get ORF Coding potential... Type: noncoding Potential: -0.706 Folding energies... MFEI: -30.570 AMFE: -0.665 GC content %: 45.985 | |
| Taestivum_Traes_1AL_A3A71720B.3 Traes_1AL_A3A71720B.3 PAC:31932758 |
Low | No | 4825 | Get ORF Coding potential... Type: noncoding Potential: -0.889 Folding energies... MFEI: -26.524 AMFE: -0.668 GC content %: 39.731 | |
| Taestivum_Traes_1AL_A3A71720B.30 Traes_1AL_A3A71720B.30 PAC:31932778 |
High | No | 2397 | Get ORF Coding potential... Type: noncoding Potential: -0.770 Folding energies... MFEI: -28.840 AMFE: -0.659 GC content %: 43.763 | |
| Taestivum_Traes_1AL_A3A71720B.31 Traes_1AL_A3A71720B.31 PAC:31932779 |
High | No | 1133 | Get ORF Coding potential... Type: noncoding Potential: -0.799 Folding energies... MFEI: -29.965 AMFE: -0.667 GC content %: 44.925 | |
| Taestivum_Traes_1AL_A3A71720B.32 Traes_1AL_A3A71720B.32 PAC:31932780 |
High | No | 1404 | Get ORF Coding potential... Type: noncoding Potential: -0.987 Folding energies... MFEI: -25.883 AMFE: -0.648 GC content %: 39.957 | |
| Taestivum_Traes_1AL_A3A71720B.4 Traes_1AL_A3A71720B.4 PAC:31932757 |
Low | No | 3594 | Get ORF Coding potential... Type: noncoding Potential: -0.783 Folding energies... MFEI: -27.457 AMFE: -0.656 GC content %: 41.875 | |
| Taestivum_Traes_1AL_A3A71720B.5 Traes_1AL_A3A71720B.5 PAC:31932752 |
Low | No | 4739 | Get ORF Coding potential... Type: noncoding Potential: -0.822 Folding energies... MFEI: -27.391 AMFE: -0.656 GC content %: 41.760 | |
| Taestivum_Traes_1AL_A3A71720B.6 Traes_1AL_A3A71720B.6 PAC:31932761 |
Low | No | 4743 | Get ORF Coding potential... Type: noncoding Potential: -0.822 Folding energies... MFEI: -27.493 AMFE: -0.657 GC content %: 41.830 | |
| Taestivum_Traes_1AL_A3A71720B.7 Traes_1AL_A3A71720B.7 PAC:31932763 |
Low | No | 5405 | Get ORF Coding potential... Type: noncoding Potential: -0.829 Folding energies... MFEI: -27.541 AMFE: -0.665 GC content %: 41.443 | |
| Taestivum_Traes_1AL_A3A71720B.8 Traes_1AL_A3A71720B.8 PAC:31932756 |
Low | No | 5613 | Get ORF Coding potential... Type: noncoding Potential: -0.891 Folding energies... MFEI: -26.994 AMFE: -0.681 GC content %: 39.640 | |
| Taestivum_Traes_1AL_A3A71720B.9 Traes_1AL_A3A71720B.9 PAC:31932764 |
Low | No | 3575 | Get ORF Coding potential... Type: noncoding Potential: -0.876 Folding energies... MFEI: -27.169 AMFE: -0.658 GC content %: 41.287 |
Matches to external databases
| Transcript alias | Database | Version | Hit | Evalue |
|---|---|---|---|---|
| Taestivum_Traes_1AL_A3A71720B.17 | RepBase (RepeatMasker) | 2014-01-31 | Jittery | |
| Taestivum_Traes_1AL_A3A71720B.12 | RepBase (RepeatMasker) | 2014-01-31 | Jittery | |
| Taestivum_Traes_1AL_A3A71720B.6 | RepBase (RepeatMasker) | 2014-01-31 | Jittery | |
| Taestivum_Traes_1AL_A3A71720B.27 | RepBase (RepeatMasker) | 2014-01-31 | Jittery | |
| Taestivum_Traes_1AL_A3A71720B.18 | RepBase (RepeatMasker) | 2014-01-31 | Jittery | |
| Taestivum_Traes_1AL_A3A71720B.13 | RepBase (RepeatMasker) | 2014-01-31 | Jittery | |
| Taestivum_Traes_1AL_A3A71720B.7 | RepBase (RepeatMasker) | 2014-01-31 | Jittery | |
| Taestivum_Traes_1AL_A3A71720B.28 | RepBase (RepeatMasker) | 2014-01-31 | hAT-N19_SBi | |
| Taestivum_Traes_1AL_A3A71720B.2 | RepBase (RepeatMasker) | 2014-01-31 | Jittery | |
| Taestivum_Traes_1AL_A3A71720B.8 | RepBase (RepeatMasker) | 2014-01-31 | Jittery |
... further results
Gene models