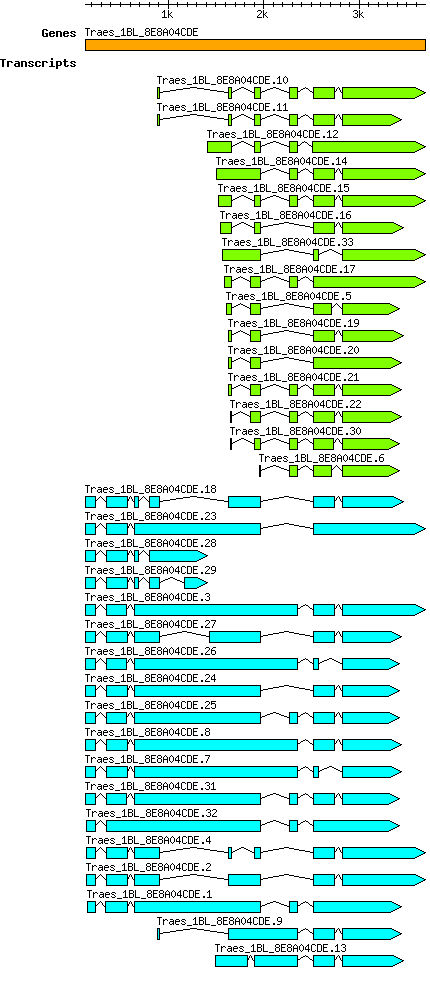
Gene
Taestivum Traes 1BL 8E8A04CDE
From GreeNC
Gene information
| Gene alias / name | Chrom. | Start | End | Source | Assembly | Species | Details
|
|---|---|---|---|---|---|---|---|
| Taestivum_Traes_1BL_8E8A04CDE Traes_1BL_8E8A04CDE |
ta_iwgsc_1bl_v1_3820857 | 131 | 3696 | Phytozome | v2.2 | Triticum_aestivum | Coding? No |
Transcript features
This table shows how a lncRNA has been classified (high/low confidence) or whether it has been classified as pre-miRNA (go to FAQ for classification criteria). Besides, it displays the lncRNA length and sequence as well as information about the ORF, the coding potential calculated by the Coding Potential Calculator (CPC), and the folding energies calculated by RNAfold.
| Transcript alias / name | Confidence | Pre-miRNA | Length | Sequence | Details
|
|---|---|---|---|---|---|
| Taestivum_Traes_1BL_8E8A04CDE.1 Traes_1BL_8E8A04CDE.1 PAC:31998777 |
Low | No | 2646 | Get ORF Coding potential... Type: noncoding Potential: -0.528 Folding energies... MFEI: -28.980 AMFE: -0.702 GC content %: 41.270 | |
| Taestivum_Traes_1BL_8E8A04CDE.10 Traes_1BL_8E8A04CDE.10 PAC:31998769 |
High | No | 1293 | Get ORF Coding potential... Type: noncoding Potential: -1.135 Folding energies... MFEI: -30.696 AMFE: -0.640 GC content %: 47.951 | |
| Taestivum_Traes_1BL_8E8A04CDE.11 Traes_1BL_8E8A04CDE.11 PAC:31998765 |
High | No | 1036 | Get ORF Coding potential... Type: noncoding Potential: -1.386 Folding energies... MFEI: -29.208 AMFE: -0.624 GC content %: 46.815 | |
| Taestivum_Traes_1BL_8E8A04CDE.12 Traes_1BL_8E8A04CDE.12 PAC:31998757 |
High | No | 1582 | Get ORF Coding potential... Type: noncoding Potential: -1.244 Folding energies... MFEI: -30.392 AMFE: -0.658 GC content %: 46.207 | |
| Taestivum_Traes_1BL_8E8A04CDE.13 Traes_1BL_8E8A04CDE.13 PAC:31998763 |
Low | No | 1643 | Get ORF Coding potential... Type: noncoding Potential: -1.302 Folding energies... MFEI: -27.553 AMFE: -0.633 GC content %: 43.518 | |
| Taestivum_Traes_1BL_8E8A04CDE.14 Traes_1BL_8E8A04CDE.14 PAC:31998758 |
High | No | 1643 | Get ORF Coding potential... Type: noncoding Potential: -1.276 Folding energies... MFEI: -29.519 AMFE: -0.649 GC content %: 45.466 | |
| Taestivum_Traes_1BL_8E8A04CDE.15 Traes_1BL_8E8A04CDE.15 PAC:31998752 |
High | No | 1375 | Get ORF Coding potential... Type: noncoding Potential: -1.141 Folding energies... MFEI: -30.596 AMFE: -0.648 GC content %: 47.200 | |
| Taestivum_Traes_1BL_8E8A04CDE.16 Traes_1BL_8E8A04CDE.16 PAC:31998771 |
High | No | 1037 | Get ORF Coding potential... Type: noncoding Potential: -1.298 Folding energies... MFEI: -29.691 AMFE: -0.630 GC content %: 47.155 | |
| Taestivum_Traes_1BL_8E8A04CDE.17 Traes_1BL_8E8A04CDE.17 PAC:31998760 |
High | No | 1443 | Get ORF Coding potential... Type: noncoding Potential: -1.145 Folding energies... MFEI: -30.416 AMFE: -0.655 GC content %: 46.431 | |
| Taestivum_Traes_1BL_8E8A04CDE.18 Traes_1BL_8E8A04CDE.18 PAC:31998753 |
Low | No | 1673 | Get ORF Coding potential... Type: noncoding Potential: -1.155 Folding energies... MFEI: -28.553 AMFE: -0.639 GC content %: 44.650 | |
| Taestivum_Traes_1BL_8E8A04CDE.19 Traes_1BL_8E8A04CDE.19 PAC:31998767 |
High | No | 1005 | Get ORF Coding potential... Type: noncoding Potential: -1.383 Folding energies... MFEI: -28.259 AMFE: -0.600 GC content %: 47.065 | |
| Taestivum_Traes_1BL_8E8A04CDE.2 Traes_1BL_8E8A04CDE.2 PAC:31998748 |
Low | No | 2010 | Get ORF Coding potential... Type: noncoding Potential: -0.505 Folding energies... MFEI: -29.338 AMFE: -0.652 GC content %: 44.975 | |
| Taestivum_Traes_1BL_8E8A04CDE.20 Traes_1BL_8E8A04CDE.20 PAC:31998764 |
High | No | 1068 | Get ORF Coding potential... Type: noncoding Potential: -1.308 Folding energies... MFEI: -28.333 AMFE: -0.614 GC content %: 46.161 | |
| Taestivum_Traes_1BL_8E8A04CDE.21 Traes_1BL_8E8A04CDE.21 PAC:31998768 |
High | No | 1067 | Get ORF Coding potential... Type: noncoding Potential: -1.388 Folding energies... MFEI: -28.754 AMFE: -0.624 GC content %: 46.111 | |
| Taestivum_Traes_1BL_8E8A04CDE.22 Traes_1BL_8E8A04CDE.22 PAC:31998761 |
High | No | 1055 | Get ORF Coding potential... Type: noncoding Potential: -1.387 Folding energies... MFEI: -29.289 AMFE: -0.632 GC content %: 46.351 | |
| Taestivum_Traes_1BL_8E8A04CDE.23 Traes_1BL_8E8A04CDE.23 PAC:31998755 |
Low | No | 2830 | Get ORF Coding potential... Type: noncoding Potential: -0.531 Folding energies... MFEI: -29.452 AMFE: -0.694 GC content %: 42.438 | |
| Taestivum_Traes_1BL_8E8A04CDE.24 Traes_1BL_8E8A04CDE.24 PAC:31998770 |
Low | No | 2475 | Get ORF Coding potential... Type: noncoding Potential: -0.502 Folding energies... MFEI: -29.115 AMFE: -0.700 GC content %: 41.616 | |
| Taestivum_Traes_1BL_8E8A04CDE.25 Traes_1BL_8E8A04CDE.25 PAC:31998772 |
Low | No | 2551 | Get ORF Coding potential... Type: noncoding Potential: -1.225 Folding energies... MFEI: -29.079 AMFE: -0.702 GC content %: 41.396 | |
| Taestivum_Traes_1BL_8E8A04CDE.26 Traes_1BL_8E8A04CDE.26 PAC:31998766 |
Low | No | 2677 | Get ORF Coding potential... Type: noncoding Potential: -1.228 Folding energies... MFEI: -28.752 AMFE: -0.706 GC content %: 40.717 | |
| Taestivum_Traes_1BL_8E8A04CDE.27 Traes_1BL_8E8A04CDE.27 PAC:31998762 |
Low | No | 1967 | Get ORF Coding potential... Type: noncoding Potential: -0.470 Folding energies... MFEI: -28.876 AMFE: -0.666 GC content %: 43.366 | |
| Taestivum_Traes_1BL_8E8A04CDE.28 Traes_1BL_8E8A04CDE.28 PAC:31998773 |
Low | No | 975 | Get ORF Coding potential... Type: noncoding Potential: -1.096 Folding energies... MFEI: -28.533 AMFE: -0.744 GC content %: 38.359 | |
| Taestivum_Traes_1BL_8E8A04CDE.29 Traes_1BL_8E8A04CDE.29 PAC:31998774 |
Low | No | 712 | Get ORF Coding potential... Type: noncoding Potential: -1.046 Folding energies... MFEI: -27.331 AMFE: -0.688 GC content %: 39.747 | |
| Taestivum_Traes_1BL_8E8A04CDE.3 Traes_1BL_8E8A04CDE.3 PAC:31998747 |
Low | No | 3128 | Get ORF Coding potential... Type: noncoding Potential: -1.237 Folding energies... MFEI: -29.092 AMFE: -0.695 GC content %: 41.880 | |
| Taestivum_Traes_1BL_8E8A04CDE.30 Traes_1BL_8E8A04CDE.30 PAC:31998775 |
High | No | 964 | Get ORF Coding potential... Type: noncoding Potential: -1.380 Folding energies... MFEI: -29.948 AMFE: -0.626 GC content %: 47.822 | |
| Taestivum_Traes_1BL_8E8A04CDE.31 Traes_1BL_8E8A04CDE.31 PAC:31998778 |
Low | No | 2547 | Get ORF Coding potential... Type: noncoding Potential: -1.225 Folding energies... MFEI: -29.109 AMFE: -0.704 GC content %: 41.343 | |
| Taestivum_Traes_1BL_8E8A04CDE.32 Traes_1BL_8E8A04CDE.32 PAC:31998776 |
Low | No | 2709 | Get ORF Coding potential... Type: noncoding Potential: -1.282 Folding energies... MFEI: -28.616 AMFE: -0.700 GC content %: 40.864 | |
| Taestivum_Traes_1BL_8E8A04CDE.33 Traes_1BL_8E8A04CDE.33 PAC:31998779 |
High | No | 1327 | Get ORF Coding potential... Type: noncoding Potential: -1.261 Folding energies... MFEI: -30.249 AMFE: -0.650 GC content %: 46.571 | |
| Taestivum_Traes_1BL_8E8A04CDE.4 Traes_1BL_8E8A04CDE.4 PAC:31998749 |
Low | No | 1765 | Get ORF Coding potential... Type: noncoding Potential: -0.491 Folding energies... MFEI: -30.164 AMFE: -0.653 GC content %: 46.176 | |
| Taestivum_Traes_1BL_8E8A04CDE.5 Traes_1BL_8E8A04CDE.5 PAC:31998751 |
High | No | 948 | Get ORF Coding potential... Type: noncoding Potential: -1.378 Folding energies... MFEI: -29.673 AMFE: -0.622 GC content %: 47.679 | |
| Taestivum_Traes_1BL_8E8A04CDE.6 Traes_1BL_8E8A04CDE.6 PAC:31998750 |
High | No | 879 | Get ORF Coding potential... Type: noncoding Potential: -1.371 Folding energies... MFEI: -30.171 AMFE: -0.630 GC content %: 47.895 | |
| Taestivum_Traes_1BL_8E8A04CDE.7 Traes_1BL_8E8A04CDE.7 PAC:31998759 |
Low | No | 2701 | Get ORF Coding potential... Type: noncoding Potential: -0.516 Folding energies... MFEI: -28.515 AMFE: -0.701 GC content %: 40.652 | |
| Taestivum_Traes_1BL_8E8A04CDE.8 Traes_1BL_8E8A04CDE.8 PAC:31998756 |
Low | No | 2878 | Get ORF Coding potential... Type: noncoding Potential: -0.535 Folding energies... MFEI: -28.256 AMFE: -0.691 GC content %: 40.862 | |
| Taestivum_Traes_1BL_8E8A04CDE.9 Traes_1BL_8E8A04CDE.9 PAC:31998754 |
Low | No | 1592 | Get ORF Coding potential... Type: noncoding Potential: -1.274 Folding energies... MFEI: -27.299 AMFE: -0.631 GC content %: 43.279 |
Matches to external databases
| Transcript alias | Database | Version | Hit | Evalue |
|---|---|---|---|---|
| Taestivum_Traes_1BL_8E8A04CDE.9 | RepBase (RepeatMasker) | 2014-01-31 | ATCopia12I | |
| Taestivum_Traes_1BL_8E8A04CDE.13 | RepBase (RepeatMasker) | 2014-01-31 | ATCopia12I | |
| Taestivum_Traes_1BL_8E8A04CDE.26 | RepBase (RepeatMasker) | 2014-01-31 | ATCopia12I | |
| Taestivum_Traes_1BL_8E8A04CDE.3 | RepBase (RepeatMasker) | 2014-01-31 | ATCopia12I | |
| Taestivum_Traes_1BL_8E8A04CDE.7 | RepBase (RepeatMasker) | 2014-01-31 | ATCopia12I | |
| Taestivum_Traes_1BL_8E8A04CDE.8 | RepBase (RepeatMasker) | 2014-01-31 | ATCopia12I |
Gene models