Gene
Taestivum Traes 2BS CDE410A7D
From GreeNC
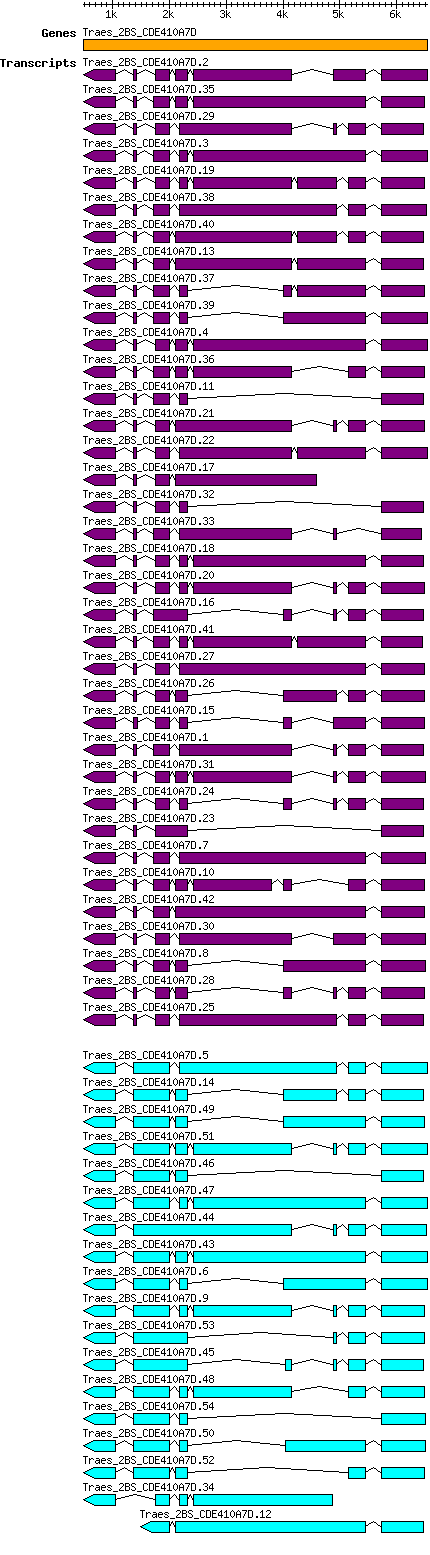
Gene information
| Gene alias / name | Chrom. | Start | End | Source | Assembly | Species | Details
|
|---|---|---|---|---|---|---|---|
| Taestivum_Traes_2BS_CDE410A7D Traes_2BS_CDE410A7D |
ta_iwgsc_2bs_v1_5233903 | 485 | 6544 | Phytozome | v2.2 | Triticum_aestivum | Coding? Yes |
Transcript features
This table shows how a lncRNA has been classified (high/low confidence) or whether it has been classified as pre-miRNA (go to FAQ for classification criteria). Besides, it displays the lncRNA length and sequence as well as information about the ORF, the coding potential calculated by the Coding Potential Calculator (CPC), and the folding energies calculated by RNAfold.
| Transcript alias / name | Confidence | Pre-miRNA | Length | Sequence | Details
|
|---|---|---|---|---|---|
| Taestivum_Traes_2BS_CDE410A7D.12 Traes_2BS_CDE410A7D.12 PAC:31810149 |
Low | No | 4611 | Get ORF Coding potential... Type: noncoding Potential: -1.102 Folding energies... MFEI: -27.662 AMFE: -0.669 GC content %: 41.336 | |
| Taestivum_Traes_2BS_CDE410A7D.14 Traes_2BS_CDE410A7D.14 PAC:31810152 |
Low | No | 3387 | Get ORF Coding potential... Type: noncoding Potential: -1.091 Folding energies... MFEI: -28.751 AMFE: -0.654 GC content %: 43.992 | |
| Taestivum_Traes_2BS_CDE410A7D.34 Traes_2BS_CDE410A7D.34 PAC:31810172 |
Low | No | 3405 | Get ORF Coding potential... Type: noncoding Potential: -1.212 Folding energies... MFEI: -25.512 AMFE: -0.671 GC content %: 38.032 | |
| Taestivum_Traes_2BS_CDE410A7D.43 Traes_2BS_CDE410A7D.43 PAC:31810182 |
Low | No | 5253 | Get ORF Coding potential... Type: noncoding Potential: -1.106 Folding energies... MFEI: -27.769 AMFE: -0.669 GC content %: 41.481 | |
| Taestivum_Traes_2BS_CDE410A7D.44 Traes_2BS_CDE410A7D.44 PAC:31810183 |
Low | No | 4496 | Get ORF Coding potential... Type: noncoding Potential: -1.101 Folding energies... MFEI: -28.514 AMFE: -0.669 GC content %: 42.593 | |
| Taestivum_Traes_2BS_CDE410A7D.45 Traes_2BS_CDE410A7D.45 PAC:31810185 |
Low | No | 2721 | Get ORF Coding potential... Type: noncoding Potential: -1.081 Folding energies... MFEI: -29.949 AMFE: -0.651 GC content %: 45.976 | |
| Taestivum_Traes_2BS_CDE410A7D.46 Traes_2BS_CDE410A7D.46 PAC:31810184 |
Low | No | 2156 | Get ORF Coding potential... Type: noncoding Potential: -1.068 Folding energies... MFEI: -30.826 AMFE: -0.644 GC content %: 47.866 | |
| Taestivum_Traes_2BS_CDE410A7D.47 Traes_2BS_CDE410A7D.47 PAC:31810187 |
Low | No | 5191 | Get ORF Coding potential... Type: noncoding Potential: -1.105 Folding energies... MFEI: -27.894 AMFE: -0.669 GC content %: 41.688 | |
| Taestivum_Traes_2BS_CDE410A7D.48 Traes_2BS_CDE410A7D.48 PAC:31810186 |
Low | No | 4138 | Get ORF Coding potential... Type: noncoding Potential: -1.098 Folding energies... MFEI: -28.830 AMFE: -0.668 GC content %: 43.137 | |
| Taestivum_Traes_2BS_CDE410A7D.49 Traes_2BS_CDE410A7D.49 PAC:31810188 |
Low | No | 3619 | Get ORF Coding potential... Type: noncoding Potential: -1.093 Folding energies... MFEI: -28.618 AMFE: -0.654 GC content %: 43.769 | |
| Taestivum_Traes_2BS_CDE410A7D.5 Traes_2BS_CDE410A7D.5 PAC:31810141 |
Low | No | 5088 | Get ORF Coding potential... Type: noncoding Potential: -1.105 Folding energies... MFEI: -27.811 AMFE: -0.664 GC content %: 41.883 | |
| Taestivum_Traes_2BS_CDE410A7D.50 Traes_2BS_CDE410A7D.50 PAC:31810189 |
Low | No | 3536 | Get ORF Coding potential... Type: noncoding Potential: -1.093 Folding energies... MFEI: -28.897 AMFE: -0.657 GC content %: 44.005 | |
| Taestivum_Traes_2BS_CDE410A7D.51 Traes_2BS_CDE410A7D.51 PAC:31810191 |
Low | No | 4307 | Get ORF Coding potential... Type: noncoding Potential: -1.100 Folding energies... MFEI: -28.563 AMFE: -0.666 GC content %: 42.884 | |
| Taestivum_Traes_2BS_CDE410A7D.52 Traes_2BS_CDE410A7D.52 PAC:31810193 |
Low | No | 2482 | Get ORF Coding potential... Type: noncoding Potential: -1.076 Folding energies... MFEI: -30.870 AMFE: -0.657 GC content %: 47.019 | |
| Taestivum_Traes_2BS_CDE410A7D.53 Traes_2BS_CDE410A7D.53 PAC:31810190 |
Low | No | 2635 | Get ORF Coding potential... Type: noncoding Potential: -1.079 Folding energies... MFEI: -30.148 AMFE: -0.652 GC content %: 46.224 | |
| Taestivum_Traes_2BS_CDE410A7D.54 Traes_2BS_CDE410A7D.54 PAC:31810192 |
Low | No | 2123 | Get ORF Coding potential... Type: noncoding Potential: -1.067 Folding energies... MFEI: -31.079 AMFE: -0.637 GC content %: 48.752 | |
| Taestivum_Traes_2BS_CDE410A7D.6 Traes_2BS_CDE410A7D.6 PAC:31810144 |
Low | No | 3606 | Get ORF Coding potential... Type: noncoding Potential: -1.093 Folding energies... MFEI: -28.760 AMFE: -0.651 GC content %: 44.204 | |
| Taestivum_Traes_2BS_CDE410A7D.9 Traes_2BS_CDE410A7D.9 PAC:31810147 |
Low | No | 4187 | Get ORF Coding potential... Type: noncoding Potential: -1.099 Folding energies... MFEI: -28.768 AMFE: -0.668 GC content %: 43.038 |
Matches to external databases
| Transcript alias | Database | Version | Hit | Evalue |
|---|---|---|---|---|
| Taestivum_Traes_2BS_CDE410A7D.47 | RepBase (RepeatMasker) | 2014-01-31 | SINE2-1_TAe | |
| Taestivum_Traes_2BS_CDE410A7D.34 | RepBase (RepeatMasker) | 2014-01-31 | SINE2-1_TAe | |
| Taestivum_Traes_2BS_CDE410A7D.52 | RepBase (RepeatMasker) | 2014-01-31 | LINE1-60_SBi | |
| Taestivum_Traes_2BS_CDE410A7D.48 | RepBase (RepeatMasker) | 2014-01-31 | SINE2-1_TAe | |
| Taestivum_Traes_2BS_CDE410A7D.43 | RepBase (RepeatMasker) | 2014-01-31 | SINE2-1_TAe | |
| Taestivum_Traes_2BS_CDE410A7D.53 | RepBase (RepeatMasker) | 2014-01-31 | LINE1-60_SBi | |
| Taestivum_Traes_2BS_CDE410A7D.49 | RepBase (RepeatMasker) | 2014-01-31 | LINE1-60_SBi | |
| Taestivum_Traes_2BS_CDE410A7D.44 | RepBase (RepeatMasker) | 2014-01-31 | SINE2-1_TAe | |
| Taestivum_Traes_2BS_CDE410A7D.54 | RepBase (RepeatMasker) | 2014-01-31 | LINE1-60_SBi | |
| Taestivum_Traes_2BS_CDE410A7D.5 | RepBase (RepeatMasker) | 2014-01-31 | SINE2-1_TAe |
... further results
Gene models