Gene
Taestivum Traes 2DL 7C079C6E6
From GreeNC
Gene information
| Gene alias / name | Chrom. | Start | End | Source | Assembly | Species | Details
|
|---|---|---|---|---|---|---|---|
| Taestivum_Traes_2DL_7C079C6E6 Traes_2DL_7C079C6E6 |
ta_iwgsc_2dl_v1_9908256 | 3335 | 9666 | Phytozome | v2.2 | Triticum_aestivum | Coding? No |
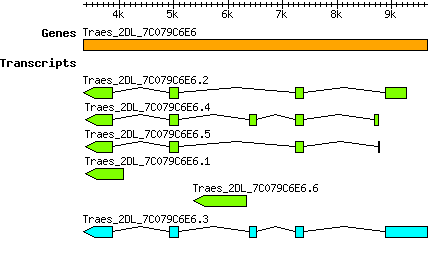
Transcript features
This table shows how a lncRNA has been classified (high/low confidence) or whether it has been classified as pre-miRNA (go to FAQ for classification criteria). Besides, it displays the lncRNA length and sequence as well as information about the ORF, the coding potential calculated by the Coding Potential Calculator (CPC), and the folding energies calculated by RNAfold.
| Transcript alias / name | Confidence | Pre-miRNA | Length | Sequence | Details
|
|---|---|---|---|---|---|
| Taestivum_Traes_2DL_7C079C6E6.1 Traes_2DL_7C079C6E6.1 PAC:31750983 |
High | No | 698 | Get ORF Coding potential... Type: noncoding Potential: -0.996 Folding energies... MFEI: -25.559 AMFE: -0.609 GC content %: 41.977 | |
| Taestivum_Traes_2DL_7C079C6E6.2 Traes_2DL_7C079C6E6.2 PAC:31750984 |
High | No | 1222 | Get ORF Coding potential... Type: noncoding Potential: -1.245 Folding energies... MFEI: -29.689 AMFE: -0.681 GC content %: 43.617 | |
| Taestivum_Traes_2DL_7C079C6E6.3 Traes_2DL_7C079C6E6.3 PAC:31750985 |
Low | Yes | 1747 | Get ORF Coding potential... Type: noncoding Potential: -1.213 Folding energies... MFEI: -28.695 AMFE: -0.682 GC content %: 42.072 | |
| Taestivum_Traes_2DL_7C079C6E6.4 Traes_2DL_7C079C6E6.4 PAC:31750987 |
High | No | 987 | Get ORF Coding potential... Type: noncoding Potential: -1.131 Folding energies... MFEI: -29.037 AMFE: -0.637 GC content %: 45.593 | |
| Taestivum_Traes_2DL_7C079C6E6.5 Traes_2DL_7C079C6E6.5 PAC:31750986 |
High | No | 824 | Get ORF Coding potential... Type: noncoding Potential: -1.205 Folding energies... MFEI: -30.413 AMFE: -0.639 GC content %: 47.573 | |
| Taestivum_Traes_2DL_7C079C6E6.6 Traes_2DL_7C079C6E6.6 PAC:31750988 |
High | No | 960 | Get ORF Coding potential... Type: noncoding Potential: -1.232 Folding energies... MFEI: -25.344 AMFE: -0.637 GC content %: 39.792 |
Matches to external databases
| Transcript alias | Database | Version | Hit | Evalue |
|---|---|---|---|---|
| Taestivum_Traes_2DL_7C079C6E6.3 | Rfam | 11.0 | MIR1122 | 1.1e-8 |
Gene models