Gene
Taestivum Traes 3B A2AECCB08
From GreeNC
Gene information
| Gene alias / name | Chrom. | Start | End | Source | Assembly | Species | Details
|
|---|---|---|---|---|---|---|---|
| Taestivum_Traes_3B_A2AECCB08 Traes_3B_A2AECCB08 |
ta_iwgsc_3b_v1_10495109 | 8 | 3777 | Phytozome | v2.2 | Triticum_aestivum | Coding? Yes |
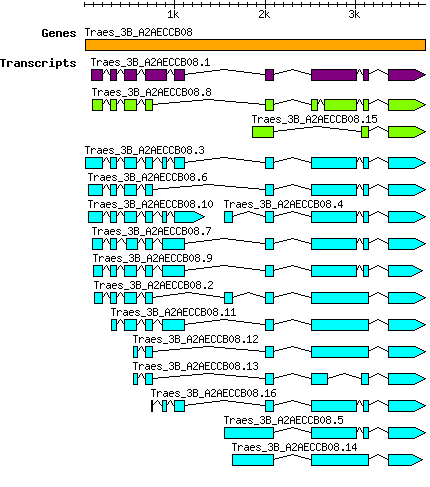
Transcript features
This table shows how a lncRNA has been classified (high/low confidence) or whether it has been classified as pre-miRNA (go to FAQ for classification criteria). Besides, it displays the lncRNA length and sequence as well as information about the ORF, the coding potential calculated by the Coding Potential Calculator (CPC), and the folding energies calculated by RNAfold.
| Transcript alias / name | Confidence | Pre-miRNA | Length | Sequence | Details
|
|---|---|---|---|---|---|
| Taestivum_Traes_3B_A2AECCB08.10 Traes_3B_A2AECCB08.10 PAC:31876764 |
Low | Yes | 808 | Get ORF Coding potential... Type: noncoding Potential: -1.147 Folding energies... MFEI: -35.111 AMFE: -0.700 GC content %: 50.124 | |
| Taestivum_Traes_3B_A2AECCB08.11 Traes_3B_A2AECCB08.11 PAC:31876765 |
Low | No | 1660 | Get ORF Coding potential... Type: noncoding Potential: -1.055 Folding energies... MFEI: -28.145 AMFE: -0.677 GC content %: 41.566 | |
| Taestivum_Traes_3B_A2AECCB08.12 Traes_3B_A2AECCB08.12 PAC:31876767 |
Low | No | 1251 | Get ORF Coding potential... Type: noncoding Potential: -1.025 Folding energies... MFEI: -26.123 AMFE: -0.640 GC content %: 40.847 | |
| Taestivum_Traes_3B_A2AECCB08.13 Traes_3B_A2AECCB08.13 PAC:31876768 |
Low | No | 875 | Get ORF Coding potential... Type: noncoding Potential: -0.975 Folding energies... MFEI: -25.269 AMFE: -0.609 GC content %: 41.486 | |
| Taestivum_Traes_3B_A2AECCB08.14 Traes_3B_A2AECCB08.14 PAC:31876769 |
Low | No | 1471 | Get ORF Coding potential... Type: noncoding Potential: -1.043 Folding energies... MFEI: -27.709 AMFE: -0.656 GC content %: 42.216 | |
| Taestivum_Traes_3B_A2AECCB08.15 Traes_3B_A2AECCB08.15 PAC:31876766 |
High | No | 723 | Get ORF Coding potential... Type: noncoding Potential: -0.941 Folding energies... MFEI: -24.219 AMFE: -0.592 GC content %: 40.941 | |
| Taestivum_Traes_3B_A2AECCB08.16 Traes_3B_A2AECCB08.16 PAC:31876770 |
Low | No | 1225 | Get ORF Coding potential... Type: noncoding Potential: -1.101 Folding energies... MFEI: -27.282 AMFE: -0.670 GC content %: 40.735 | |
| Taestivum_Traes_3B_A2AECCB08.2 Traes_3B_A2AECCB08.2 PAC:31876755 |
Low | No | 1598 | Get ORF Coding potential... Type: noncoding Potential: -1.126 Folding energies... MFEI: -28.717 AMFE: -0.651 GC content %: 44.118 | |
| Taestivum_Traes_3B_A2AECCB08.3 Traes_3B_A2AECCB08.3 PAC:31876763 |
Low | No | 1677 | Get ORF Coding potential... Type: noncoding Potential: -0.757 Folding energies... MFEI: -31.765 AMFE: -0.695 GC content %: 45.736 | |
| Taestivum_Traes_3B_A2AECCB08.4 Traes_3B_A2AECCB08.4 PAC:31876760 |
Low | No | 1157 | Get ORF Coding potential... Type: noncoding Potential: -0.720 Folding energies... MFEI: -24.952 AMFE: -0.614 GC content %: 40.622 | |
| Taestivum_Traes_3B_A2AECCB08.5 Traes_3B_A2AECCB08.5 PAC:31876759 |
Low | No | 1515 | Get ORF Coding potential... Type: noncoding Potential: -0.748 Folding energies... MFEI: -27.597 AMFE: -0.652 GC content %: 42.310 | |
| Taestivum_Traes_3B_A2AECCB08.6 Traes_3B_A2AECCB08.6 PAC:31876762 |
Low | No | 1494 | Get ORF Coding potential... Type: noncoding Potential: -0.746 Folding energies... MFEI: -30.120 AMFE: -0.664 GC content %: 45.382 | |
| Taestivum_Traes_3B_A2AECCB08.7 Traes_3B_A2AECCB08.7 PAC:31876757 |
Low | No | 1677 | Get ORF Coding potential... Type: noncoding Potential: -0.757 Folding energies... MFEI: -29.797 AMFE: -0.673 GC content %: 44.305 | |
| Taestivum_Traes_3B_A2AECCB08.8 Traes_3B_A2AECCB08.8 PAC:31876758 |
High | No | 1370 | Get ORF Coding potential... Type: noncoding Potential: -0.738 Folding energies... MFEI: -30.058 AMFE: -0.664 GC content %: 45.255 | |
| Taestivum_Traes_3B_A2AECCB08.9 Traes_3B_A2AECCB08.9 PAC:31876761 |
Low | No | 1636 | Get ORF Coding potential... Type: noncoding Potential: -0.755 Folding energies... MFEI: -29.939 AMFE: -0.677 GC content %: 44.193 |
Matches to external databases
| Transcript alias | Database | Version | Hit | Evalue |
|---|---|---|---|---|
| Taestivum_Traes_3B_A2AECCB08.13 | RepBase (RepeatMasker) | 2014-01-31 | hAT-1_HV | |
| Taestivum_Traes_3B_A2AECCB08.5 | RepBase (RepeatMasker) | 2014-01-31 | SINE2-1_TAe | |
| Taestivum_Traes_3B_A2AECCB08.14 | RepBase (RepeatMasker) | 2014-01-31 | SINE2-1_TAe | |
| Taestivum_Traes_3B_A2AECCB08.6 | RepBase (RepeatMasker) | 2014-01-31 | hAT-1_HV | |
| Taestivum_Traes_3B_A2AECCB08.16 | RepBase (RepeatMasker) | 2014-01-31 | hAT-1_HV | |
| Taestivum_Traes_3B_A2AECCB08.7 | RepBase (RepeatMasker) | 2014-01-31 | hAT-1_HV | |
| Taestivum_Traes_3B_A2AECCB08.2 | RepBase (RepeatMasker) | 2014-01-31 | hAT-1_HV | |
| Taestivum_Traes_3B_A2AECCB08.11 | RepBase (RepeatMasker) | 2014-01-31 | hAT-1_HV | |
| Taestivum_Traes_3B_A2AECCB08.9 | RepBase (RepeatMasker) | 2014-01-31 | hAT-1_HV | |
| Taestivum_Traes_3B_A2AECCB08.3 | RepBase (RepeatMasker) | 2014-01-31 | hAT-1_HV |
... further results
Gene models