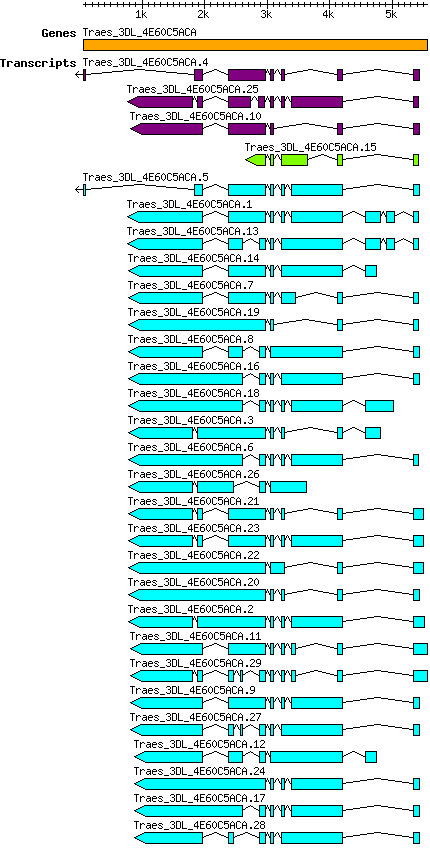
Gene
Taestivum Traes 3DL 4E60C5ACA
From GreeNC
Gene information
| Gene alias / name | Chrom. | Start | End | Source | Assembly | Species | Details
|
|---|---|---|---|---|---|---|---|
| Taestivum_Traes_3DL_4E60C5ACA Traes_3DL_4E60C5ACA |
ta_iwgsc_3dl_v1_1707087 | 63 | 5562 | Phytozome | v2.2 | Triticum_aestivum | Coding? Yes |
Transcript features
This table shows how a lncRNA has been classified (high/low confidence) or whether it has been classified as pre-miRNA (go to FAQ for classification criteria). Besides, it displays the lncRNA length and sequence as well as information about the ORF, the coding potential calculated by the Coding Potential Calculator (CPC), and the folding energies calculated by RNAfold.
| Transcript alias / name | Confidence | Pre-miRNA | Length | Sequence | Details
|
|---|---|---|---|---|---|
| Taestivum_Traes_3DL_4E60C5ACA.1 Traes_3DL_4E60C5ACA.1 PAC:31871715 |
Low | Yes | 3156 | Get ORF Coding potential... Type: noncoding Potential: -0.086 Folding energies... MFEI: -25.545 AMFE: -0.681 GC content %: 37.484 | |
| Taestivum_Traes_3DL_4E60C5ACA.11 Traes_3DL_4E60C5ACA.11 PAC:31871716 |
Low | Yes | 2210 | Get ORF Coding potential... Type: noncoding Potential: -0.010 Folding energies... MFEI: -26.683 AMFE: -0.710 GC content %: 37.602 | |
| Taestivum_Traes_3DL_4E60C5ACA.12 Traes_3DL_4E60C5ACA.12 PAC:31871711 |
Low | Yes | 2732 | Get ORF Coding potential... Type: noncoding Potential: -0.064 Folding energies... MFEI: -24.176 AMFE: -0.666 GC content %: 36.274 | |
| Taestivum_Traes_3DL_4E60C5ACA.13 Traes_3DL_4E60C5ACA.13 PAC:31871718 |
Low | Yes | 2999 | Get ORF Coding potential... Type: noncoding Potential: -0.073 Folding energies... MFEI: -24.985 AMFE: -0.671 GC content %: 37.212 | |
| Taestivum_Traes_3DL_4E60C5ACA.14 Traes_3DL_4E60C5ACA.14 PAC:31871710 |
Low | Yes | 2983 | Get ORF Coding potential... Type: noncoding Potential: -0.072 Folding energies... MFEI: -24.740 AMFE: -0.679 GC content %: 36.440 | |
| Taestivum_Traes_3DL_4E60C5ACA.15 Traes_3DL_4E60C5ACA.15 PAC:31871719 |
High | No | 962 | Get ORF Coding potential... Type: noncoding Potential: -0.964 Folding energies... MFEI: -25.104 AMFE: -0.656 GC content %: 38.254 | |
| Taestivum_Traes_3DL_4E60C5ACA.16 Traes_3DL_4E60C5ACA.16 PAC:31871721 |
Low | Yes | 3044 | Get ORF Coding potential... Type: noncoding Potential: -0.237 Folding energies... MFEI: -24.905 AMFE: -0.692 GC content %: 36.005 | |
| Taestivum_Traes_3DL_4E60C5ACA.17 Traes_3DL_4E60C5ACA.17 PAC:31871722 |
Low | Yes | 2832 | Get ORF Coding potential... Type: noncoding Potential: -0.231 Folding energies... MFEI: -25.120 AMFE: -0.689 GC content %: 36.476 | |
| Taestivum_Traes_3DL_4E60C5ACA.18 Traes_3DL_4E60C5ACA.18 PAC:31871720 |
Low | Yes | 3286 | Get ORF Coding potential... Type: noncoding Potential: -0.251 Folding energies... MFEI: -25.313 AMFE: -0.680 GC content %: 37.249 | |
| Taestivum_Traes_3DL_4E60C5ACA.19 Traes_3DL_4E60C5ACA.19 PAC:31871725 |
Low | Yes | 2406 | Get ORF Coding potential... Type: noncoding Potential: -0.180 Folding energies... MFEI: -26.297 AMFE: -0.725 GC content %: 36.284 | |
| Taestivum_Traes_3DL_4E60C5ACA.2 Traes_3DL_4E60C5ACA.2 PAC:31871706 |
Low | Yes | 3198 | Get ORF Coding potential... Type: noncoding Potential: -0.272 Folding energies... MFEI: -25.600 AMFE: -0.694 GC content %: 36.867 | |
| Taestivum_Traes_3DL_4E60C5ACA.20 Traes_3DL_4E60C5ACA.20 PAC:31871729 |
Low | Yes | 2457 | Get ORF Coding potential... Type: noncoding Potential: -0.232 Folding energies... MFEI: -26.435 AMFE: -0.727 GC content %: 36.345 | |
| Taestivum_Traes_3DL_4E60C5ACA.21 Traes_3DL_4E60C5ACA.21 PAC:31871726 |
Low | Yes | 2031 | Get ORF Coding potential... Type: noncoding Potential: -0.120 Folding energies... MFEI: -26.199 AMFE: -0.713 GC content %: 36.731 | |
| Taestivum_Traes_3DL_4E60C5ACA.22 Traes_3DL_4E60C5ACA.22 PAC:31871727 |
Low | Yes | 2644 | Get ORF Coding potential... Type: noncoding Potential: -0.283 Folding energies... MFEI: -26.524 AMFE: -0.728 GC content %: 36.422 | |
| Taestivum_Traes_3DL_4E60C5ACA.23 Traes_3DL_4E60C5ACA.23 PAC:31871728 |
Low | Yes | 2764 | Get ORF Coding potential... Type: noncoding Potential: -0.183 Folding energies... MFEI: -25.239 AMFE: -0.689 GC content %: 36.650 | |
| Taestivum_Traes_3DL_4E60C5ACA.24 Traes_3DL_4E60C5ACA.24 PAC:31871724 |
Low | Yes | 3113 | Get ORF Coding potential... Type: noncoding Potential: -0.270 Folding energies... MFEI: -25.365 AMFE: -0.697 GC content %: 36.396 | |
| Taestivum_Traes_3DL_4E60C5ACA.26 Traes_3DL_4E60C5ACA.26 PAC:31871730 |
Low | Yes | 2259 | Get ORF Coding potential... Type: noncoding Potential: -1.298 Folding energies... MFEI: -24.533 AMFE: -0.693 GC content %: 35.414 | |
| Taestivum_Traes_3DL_4E60C5ACA.27 Traes_3DL_4E60C5ACA.27 PAC:31871732 |
Low | Yes | 2497 | Get ORF Coding potential... Type: noncoding Potential: -1.095 Folding energies... MFEI: -24.021 AMFE: -0.674 GC content %: 35.643 | |
| Taestivum_Traes_3DL_4E60C5ACA.28 Traes_3DL_4E60C5ACA.28 PAC:31871731 |
Low | Yes | 2388 | Get ORF Coding potential... Type: noncoding Potential: -1.166 Folding energies... MFEI: -24.083 AMFE: -0.680 GC content %: 35.427 | |
| Taestivum_Traes_3DL_4E60C5ACA.29 Traes_3DL_4E60C5ACA.29 PAC:31871733 |
Low | Yes | 1750 | Get ORF Coding potential... Type: noncoding Potential: -1.046 Folding energies... MFEI: -25.897 AMFE: -0.684 GC content %: 37.886 | |
| Taestivum_Traes_3DL_4E60C5ACA.3 Traes_3DL_4E60C5ACA.3 PAC:31871705 |
Low | Yes | 2522 | Get ORF Coding potential... Type: noncoding Potential: -0.210 Folding energies... MFEI: -26.356 AMFE: -0.706 GC content %: 37.351 | |
| Taestivum_Traes_3DL_4E60C5ACA.5 Traes_3DL_4E60C5ACA.5 PAC:31871707 |
Low | No | 1739 | Get ORF Coding potential... Type: noncoding Potential: -0.082 Folding energies... MFEI: -25.888 AMFE: -0.669 GC content %: 38.700 | |
| Taestivum_Traes_3DL_4E60C5ACA.6 Traes_3DL_4E60C5ACA.6 PAC:31871709 |
Low | Yes | 2922 | Get ORF Coding potential... Type: noncoding Potential: -0.211 Folding energies... MFEI: -25.212 AMFE: -0.692 GC content %: 36.413 | |
| Taestivum_Traes_3DL_4E60C5ACA.7 Traes_3DL_4E60C5ACA.7 PAC:31871712 |
Low | Yes | 2221 | Get ORF Coding potential... Type: noncoding Potential: -0.011 Folding energies... MFEI: -25.552 AMFE: -0.709 GC content %: 36.020 | |
| Taestivum_Traes_3DL_4E60C5ACA.8 Traes_3DL_4E60C5ACA.8 PAC:31871713 |
Low | Yes | 2752 | Get ORF Coding potential... Type: noncoding Potential: -0.065 Folding energies... MFEI: -24.375 AMFE: -0.683 GC content %: 35.683 | |
| Taestivum_Traes_3DL_4E60C5ACA.9 Traes_3DL_4E60C5ACA.9 PAC:31871717 |
Low | Yes | 2760 | Get ORF Coding potential... Type: noncoding Potential: -0.065 Folding energies... MFEI: -24.888 AMFE: -0.687 GC content %: 36.232 |
Matches to external databases
| Transcript alias | Database | Version | Hit | Evalue |
|---|---|---|---|---|
| Taestivum_Traes_3DL_4E60C5ACA.18 | Rfam | 11.0 | MIR1122 | 4.1e-5 |
| Taestivum_Traes_3DL_4E60C5ACA.9 | Rfam | 11.0 | MIR1122 | 3.4e-5 |
| Taestivum_Traes_3DL_4E60C5ACA.26 | RepBase (RepeatMasker) | 2014-01-31 | ATHOS_HV | |
| Taestivum_Traes_3DL_4E60C5ACA.12 | Rfam | 11.0 | MIR1122 | 3.4e-5 |
| Taestivum_Traes_3DL_4E60C5ACA.29 | Rfam | 11.0 | MIR1122 | 2.2e-5 |
| Taestivum_Traes_3DL_4E60C5ACA.20 | RepBase (RepeatMasker) | 2014-01-31 | ATHOS_HV | |
| Taestivum_Traes_3DL_4E60C5ACA.23 | Rfam | 11.0 | MIR1122 | 3.4e-5 |
| Taestivum_Traes_3DL_4E60C5ACA.16 | RepBase (RepeatMasker) | 2014-01-31 | ATHOS_HV | |
| Taestivum_Traes_3DL_4E60C5ACA.7 | RepBase (RepeatMasker) | 2014-01-31 | ATHOS_HV | |
| Taestivum_Traes_3DL_4E60C5ACA.19 | Rfam | 11.0 | MIR1122 | 3.0e-5 |
... further results
Gene models